
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 20,467,169 – 20,467,298 |
| Length | 129 |
| Max. P | 0.980187 |

| Location | 20,467,169 – 20,467,274 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.75 |
| Mean single sequence MFE | -24.65 |
| Consensus MFE | -17.81 |
| Energy contribution | -17.12 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.16 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.760276 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20467169 105 + 27905053 AUAA---C-------GA-----AUCUCACUUGAAAUAUGUGUGCUCGAAAAUGUUGCGAUCCUGCAGGAACUGUCGAUUGUCCUGCCAAAUCCAAUCCUCGAGCUCCUUGUUCAACAAUG ....---.-------..-----.......(((((....(.(.((((((....((((.(((...((((((.(........)))))))...)))))))..)))))).))...)))))..... ( -25.70) >DroVir_CAF1 18901 108 + 1 AGAAUUUU------------UAUACCCACUUGAAGUAGGUGUGCUCGAAGAUAUUUCGAUCUUGCAAGAACUGUCGGUUAUCCUGCCAAAUCCAAUCUUCCAGCUCCUUACUCAACAAUG ........------------.........((((.(((((.(.(((.((((((.......(((....)))......(((......))).......)))))).))).))))))))))..... ( -22.00) >DroGri_CAF1 23154 120 + 1 GAGAUUGUUUUUGCUUGUUGUCUGCUCACUUGAAGUAGGUGUGCUCGAAGAUGUUGCGAUCUUGCAGGAACUGUCUAUUGUCCUGCCAUAUCCAAUCCUCUAGCUCCUUGCUCAACAAUG ...((((((...((..(..((..((.((((((...)))))).))....(((.((((.(((...((((((.(........)))))))...)))))))..))).))..)..))..)))))). ( -27.80) >DroWil_CAF1 19500 97 + 1 -----------------------GCUUACUUGAAAUAGGUGUGCUCGAAGAUAUUUCGAUCUUGGAGGAAUUGUCGAUUGUCCUGCCAGAUCCAAUCUUCAAGCUCUUUAUUCAAAAAUG -----------------------......(((((..(((...(((.((((((.....(((((.((((((...........)))).)))))))..)))))).))).)))..)))))..... ( -23.80) >DroMoj_CAF1 19966 108 + 1 AUAAAUAU------------UAAUCUCACUUAAAGUAUGUGUGCUCGAAGAUAUUUCGAUCUUGCAAGAACUGUCUGUUGUCUUGCCAAAUCCAAUCUUCCAGUUCCUUGCUUAACAAAG ........------------............(((((.(.(.(((.((((((.....(((...((((((((.....))..))))))...)))..)))))).))).)).)))))....... ( -18.80) >DroAna_CAF1 17433 99 + 1 GU---------------------ACUUACUUGAAAUAAGUGUGCUCGAAGAUGUUGCGAUCUUGCAGGAACUGCCUGUUGUCCUGCCAGAUCCAAUCCUCCAGCUCCUUGUUCAACAAUG ..---------------------......(((((.((((.(.(((.((.(((..((.(((((.((((((((.....))..)))))).)))))))))).)).))).))))))))))..... ( -29.80) >consensus AUAA___U_______________ACUCACUUGAAAUAGGUGUGCUCGAAGAUAUUGCGAUCUUGCAGGAACUGUCGAUUGUCCUGCCAAAUCCAAUCCUCCAGCUCCUUGCUCAACAAUG .............................((((.(((((.(.(((.((.(((.....(((...((((((.(........)))))))...)))..))).)).))).))))))))))..... (-17.81 = -17.12 + -0.69)



| Location | 20,467,169 – 20,467,274 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.75 |
| Mean single sequence MFE | -29.49 |
| Consensus MFE | -22.91 |
| Energy contribution | -21.83 |
| Covariance contribution | -1.08 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.980187 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20467169 105 - 27905053 CAUUGUUGAACAAGGAGCUCGAGGAUUGGAUUUGGCAGGACAAUCGACAGUUCCUGCAGGAUCGCAACAUUUUCGAGCACACAUAUUUCAAGUGAGAU-----UC-------G---UUAU ........(((.....((((((((((((((((..(((((((........).))))))..)))).))..))))))))))......(((((....)))))-----..-------)---)).. ( -30.80) >DroVir_CAF1 18901 108 - 1 CAUUGUUGAGUAAGGAGCUGGAAGAUUGGAUUUGGCAGGAUAACCGACAGUUCUUGCAAGAUCGAAAUAUCUUCGAGCACACCUACUUCAAGUGGGUAUA------------AAAAUUCU .......((((.....(((.(((((((.(((((.((((((...........)))))).))))).)...)))))).)))..((((((.....))))))...------------...)))). ( -32.40) >DroGri_CAF1 23154 120 - 1 CAUUGUUGAGCAAGGAGCUAGAGGAUUGGAUAUGGCAGGACAAUAGACAGUUCCUGCAAGAUCGCAACAUCUUCGAGCACACCUACUUCAAGUGAGCAGACAACAAGCAAAAACAAUCUC ..((((((.((.....(((.(((((((((((...(((((((........).))))))...))).))..)))))).))).(((.........))).))...)))))).............. ( -30.60) >DroWil_CAF1 19500 97 - 1 CAUUUUUGAAUAAAGAGCUUGAAGAUUGGAUCUGGCAGGACAAUCGACAAUUCCUCCAAGAUCGAAAUAUCUUCGAGCACACCUAUUUCAAGUAAGC----------------------- ....((((((......(((((((((((.(((((((.((((...........)))))).))))).)...))))))))))........)))))).....----------------------- ( -26.84) >DroMoj_CAF1 19966 108 - 1 CUUUGUUAAGCAAGGAACUGGAAGAUUGGAUUUGGCAAGACAACAGACAGUUCUUGCAAGAUCGAAAUAUCUUCGAGCACACAUACUUUAAGUGAGAUUA------------AUAUUUAU (((((.....)))))..((.(((((((.(((((.(((((((........).)))))).))))).)...)))))).))..(((.........)))......------------........ ( -23.60) >DroAna_CAF1 17433 99 - 1 CAUUGUUGAACAAGGAGCUGGAGGAUUGGAUCUGGCAGGACAACAGGCAGUUCCUGCAAGAUCGCAACAUCUUCGAGCACACUUAUUUCAAGUAAGU---------------------AC ................(((.(((((((((((((.(((((((........).)))))).))))).))..)))))).)))..((((((.....))))))---------------------.. ( -32.70) >consensus CAUUGUUGAACAAGGAGCUGGAAGAUUGGAUUUGGCAGGACAACAGACAGUUCCUGCAAGAUCGAAACAUCUUCGAGCACACCUACUUCAAGUGAGA_______________A___UUAU .....(((((....(.(((.((((((..(((((.((((((...........)))))).))))).....)))))).))).)......)))))............................. (-22.91 = -21.83 + -1.08)
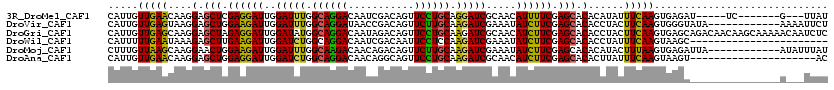


| Location | 20,467,194 – 20,467,298 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.07 |
| Mean single sequence MFE | -33.97 |
| Consensus MFE | -22.15 |
| Energy contribution | -21.99 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.37 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.967811 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20467194 104 - 27905053 -------C---------CCAGAAGCUACAACUAUUUCGUCCAUUGUUGAACAAGGAGCUCGAGGAUUGGAUUUGGCAGGACAAUCGACAGUUCCUGCAGGAUCGCAACAUUUUCGAGCAC -------.---------.....................(((.(((.....))))))((((((((((((((((..(((((((........).))))))..)))).))..)))))))))).. ( -32.10) >DroVir_CAF1 18929 120 - 1 GUGGAAGCAAAAGCGUCUAAUAAGCUACAACAGUUUCGUCCAUUGUUGAGUAAGGAGCUGGAAGAUUGGAUUUGGCAGGAUAACCGACAGUUCUUGCAAGAUCGAAAUAUCUUCGAGCAC .((((.((....)).))))....(((.(((((((.......)))))))))).....(((.(((((((.(((((.((((((...........)))))).))))).)...)))))).))).. ( -34.90) >DroGri_CAF1 23194 120 - 1 GCGGAAGCCAAAGUGUCCACAAAGCCACAACAGUUUCGUCCAUUGUUGAGCAAGGAGCUAGAGGAUUGGAUAUGGCAGGACAAUAGACAGUUCCUGCAAGAUCGCAACAUCUUCGAGCAC (((((.((....)).))).........(((((((.......))))))).)).....(((.(((((((((((...(((((((........).))))))...))).))..)))))).))).. ( -34.70) >DroWil_CAF1 19517 104 - 1 -------C---------UCAUAAAUUGCAGCUAUUUCGUCCAUUUUUGAAUAAAGAGCUUGAAGAUUGGAUCUGGCAGGACAAUCGACAAUUCCUCCAAGAUCGAAAUAUCUUCGAGCAC -------.---------.............((..((((........))))...)).(((((((((((.(((((((.((((...........)))))).))))).)...)))))))))).. ( -26.00) >DroAna_CAF1 17452 104 - 1 -------C---------CCAGAAGUUGCUACUAUUUCGUCCAUUGUUGAACAAGGAGCUGGAGGAUUGGAUCUGGCAGGACAACAGGCAGUUCCUGCAAGAUCGCAACAUCUUCGAGCAC -------.---------...(((((.......))))).(((.(((.....))))))(((.(((((((((((((.(((((((........).)))))).))))).))..)))))).))).. ( -35.20) >DroPer_CAF1 17635 110 - 1 -------C---UUCGUCCUUCAAGCUGCAGCUUCUUCGUCCUCUGCUGAACAAAGAGCUGGAGGACUGGAUCUGGCAGGACAACCGCCAGUUCCUGCAGGAGCGCAACAUCUUCGACCAC -------.---...(((........(((.((((((.(((((((((((........))).))))))).(((.(((((.((....))))))).))).).)))))))))........)))... ( -40.89) >consensus _______C_________CCAGAAGCUACAACUAUUUCGUCCAUUGUUGAACAAGGAGCUGGAGGAUUGGAUCUGGCAGGACAACCGACAGUUCCUGCAAGAUCGCAACAUCUUCGAGCAC .......................(((.((((.............))))))).....(((.((((((..(((((.((((((...........)))))).))))).....)))))).))).. (-22.15 = -21.99 + -0.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:51:01 2006