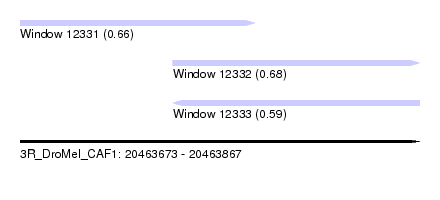
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 20,463,673 – 20,463,867 |
| Length | 194 |
| Max. P | 0.680156 |

| Location | 20,463,673 – 20,463,787 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.62 |
| Mean single sequence MFE | -48.13 |
| Consensus MFE | -28.71 |
| Energy contribution | -29.22 |
| Covariance contribution | 0.51 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.661264 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
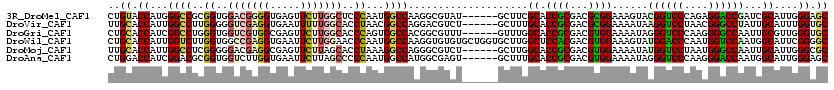
>3R_DroMel_CAF1 20463673 114 + 27905053 CUGUACCAUGGGCCGCGGUGGACGGGGUGAGUUCUUGGCUCCCAAUGGCCAAGGCGUAU------GCUUCGCACCGCGACGCGGAAAGUACGGUCCCAGAGGACCGAUCGCAUUGGGAGC ((((.((((.(....).))))))))............((((((((((((....))((((------..(((((........)))))..))))(((((....))))).....)))))))))) ( -47.20) >DroVir_CAF1 14552 114 + 1 UUGCACCAUUGGCCUUGGGGGUCGAGGUGAAUUUUUGGCACCUAACGGCCAGGACGUCU------GCUUUGCACCGCGACGCGGAAAAUAAGGUCCUAACGGGCCUAUUGCAUUUGGUGC ..(((((((((((((((((.(((((((.....))))))).))))).))))))....(((------((.((((...)))).))))).....((((((....))))))........)))))) ( -50.30) >DroGri_CAF1 18144 114 + 1 CUGCACCAUCGGCCUGGGUGGUCGUGGCGAGUUCUUGGCACCCAGUGGCCACGGCGUUU------GUUUGGCACCGCGACGUGGAAAAUAGGGUCCCAAGGGGCCAAUUGCGUUGGGUGC ..(((((..(((((.....)))))((((...(((((((.((((.((..(((((.(((.(------(.....))..))).)))))...)).)))).))))))))))).........))))) ( -47.60) >DroWil_CAF1 16031 120 + 1 CUGCACCAUUGGUCUUGGUGGCCGAGGUGAAUUCUUGGAACCCAAUGGCCAAGGUGUGUGCUGGUGCUUGGCUCCACGACGUGGAAAGUAUGGACCCAAUGGUCCAAUGGCAUUCGGGGC ..(((((.((((((((((...((((((.....))))))...)))).)))))))))))((.((((((((....(((((...))))).....((((((....))))))..)))).)))).)) ( -50.60) >DroMoj_CAF1 16519 114 + 1 UUGCACCAUUGGCCUCGGGGGACGAGGCGAGUUCUUAGCACCUAAAGGCCAGGGCGUCU------GCUUGGCACCGCGACGUGGAAAAUAUGGUCCUAAUGGGCCAAUUGCAUUGGGCGC ..((.(((((.((((((.....)))))).))).....(((.......((((((......------.)))))).((((...))))......((((((....))))))..)))...)))).. ( -42.40) >DroAna_CAF1 13275 114 + 1 CUGGACCAUCGGACGCGGUGGUCUUGGUGAAUUCUUAGCCCCCAAUGGCCAUGGCGAGU------GCUUUGCACCGCGACGUGGAAAAUAGGGUCCCAAGGGACCAAUGGCAUUGGGAGC (.(((((((((....))))))))).)...........((.(((((((.((((.(((.((------((...)))))))..............(((((....))))).))))))))))).)) ( -50.70) >consensus CUGCACCAUUGGCCUCGGUGGUCGAGGUGAAUUCUUGGCACCCAAUGGCCAAGGCGUCU______GCUUGGCACCGCGACGUGGAAAAUAGGGUCCCAAGGGGCCAAUUGCAUUGGGAGC ..((.((...((((..((..(((((((.....)))))))..))...))))....................((.((((...))))......((((((....))))))...))....)).)) (-28.71 = -29.22 + 0.51)
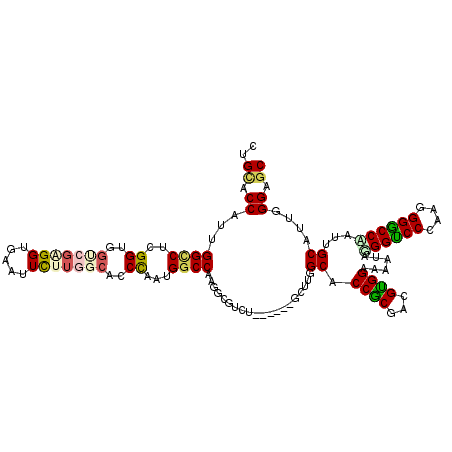

| Location | 20,463,747 – 20,463,867 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.83 |
| Mean single sequence MFE | -43.02 |
| Consensus MFE | -31.76 |
| Energy contribution | -32.43 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.680156 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20463747 120 + 27905053 GCGGAAAGUACGGUCCCAGAGGACCGAUCGCAUUGGGAGCAUUCGCCACGGACAGAUGACAGUUGAUAAGAAUGGGCGCCAGAAUACUAAUGGUUUCCAGGCUGGGAUUGCGCACCAGCU ((((......((((((....)))))).)))).((((..(((.((.((...(((........)))....((..((((.((((.........)))).))))..)))))).)))...)))).. ( -36.30) >DroVir_CAF1 14626 120 + 1 GCGGAAAAUAAGGUCCUAACGGGCCUAUUGCAUUUGGUGCAUUGGCCACGGACAAAUGGCAGUUGAUCAGAAUGGGCGCCAGAAUGAUGGUGGCCUCCAGGCUCGUGUUGCGCACUAGCU ..(((.....((((((....))))))..(((((...)))))..((((((.(.((..((((.(((.((....)).)))))))...)).).))))))))).((((.(((.....))).)))) ( -45.80) >DroGri_CAF1 18218 120 + 1 GUGGAAAAUAGGGUCCCAAGGGGCCAAUUGCGUUGGGUGCAUUGGCCACAGACAAAUGGCAGUUGAUUAAAAUGGGCGCCAAAAUGCUGGUGGCCUCCAGGCUGGUGUUGCGCACCAGCU .((((......(((((....)))))...((((.....))))..((((((((.((..((((.(((.((....)).)))))))...)))).))))))))))((((((((.....)))))))) ( -49.50) >DroWil_CAF1 16111 120 + 1 GUGGAAAGUAUGGACCCAAUGGUCCAAUGGCAUUCGGGGCAUUGGCCACUGACAAAUGGCAGUUUAUCAGAAUGGGGGCUAAAAUGCUAGUGGCUUCCAGACUUGUGUUUCGCACCAGCU ((((((.((.((((((....))))))...)).)))(..((((.((.(.((((.((((....)))).))))..(((((((((.........)))))))))).)).))))..).)))..... ( -38.10) >DroMoj_CAF1 16593 120 + 1 GUGGAAAAUAUGGUCCUAAUGGGCCAAUUGCAUUGGGCGCAUUGGCCACUGAUAAAUGGCAGUUGAUCAAAAUGGGCGCCAGAAUAGUGGUGGCCUCCAGGCUGGUGUUGCGCACCAGCU .((((.....((((((....))))))..(((.......)))..((((((((.....((((.(((.((....)).)))))))......))))))))))))((((((((.....)))))))) ( -50.00) >DroAna_CAF1 13349 120 + 1 GUGGAAAAUAGGGUCCCAAGGGACCAAUGGCAUUGGGAGCGUUCGCGACAGAAAGAUGACAAUUGAUCAGAAUGGGUGCCAAAAUGCUGGUGGCUUCUAGACUGGUGUUGCGCACCAGCU ...........(((((....)))))..(((((((.(..((....))..).((..(((....)))..))......)))))))....(((((((((.((......))....)).))))))). ( -38.40) >consensus GUGGAAAAUAGGGUCCCAAGGGGCCAAUUGCAUUGGGAGCAUUGGCCACAGACAAAUGGCAGUUGAUCAGAAUGGGCGCCAAAAUGCUGGUGGCCUCCAGGCUGGUGUUGCGCACCAGCU ((((..(((.((((((....))))))...((.......)))))..)))).......................(((..((((.........))))..)))((((((((.....)))))))) (-31.76 = -32.43 + 0.67)

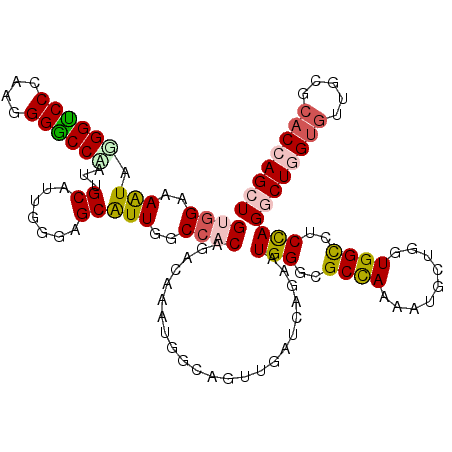
| Location | 20,463,747 – 20,463,867 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.83 |
| Mean single sequence MFE | -35.71 |
| Consensus MFE | -24.22 |
| Energy contribution | -24.53 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.34 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.586199 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20463747 120 - 27905053 AGCUGGUGCGCAAUCCCAGCCUGGAAACCAUUAGUAUUCUGGCGCCCAUUCUUAUCAACUGUCAUCUGUCCGUGGCGAAUGCUCCCAAUGCGAUCGGUCCUCUGGGACCGUACUUUCCGC .(((((.........)))))..((((((((((((((((((((...)))............(((((......))))))))))))...)))).)..((((((....))))))...))))).. ( -33.40) >DroVir_CAF1 14626 120 - 1 AGCUAGUGCGCAACACGAGCCUGGAGGCCACCAUCAUUCUGGCGCCCAUUCUGAUCAACUGCCAUUUGUCCGUGGCCAAUGCACCAAAUGCAAUAGGCCCGUUAGGACCUUAUUUUCCGC .((....))((...(((.(((((..((((((...((...(((((...............)))))..))...))))))..((((.....)))).))))).)))..(((........))))) ( -35.16) >DroGri_CAF1 18218 120 - 1 AGCUGGUGCGCAACACCAGCCUGGAGGCCACCAGCAUUUUGGCGCCCAUUUUAAUCAACUGCCAUUUGUCUGUGGCCAAUGCACCCAACGCAAUUGGCCCCUUGGGACCCUAUUUUCCAC .(((((((.....))))))).((((((((((..(((...(((((...............)))))..)))..))))))......(((((.((.....))...))))).........)))). ( -42.86) >DroWil_CAF1 16111 120 - 1 AGCUGGUGCGAAACACAAGUCUGGAAGCCACUAGCAUUUUAGCCCCCAUUCUGAUAAACUGCCAUUUGUCAGUGGCCAAUGCCCCGAAUGCCAUUGGACCAUUGGGUCCAUACUUUCCAC .((((((((....((......))...).)))))))......(..((((..((((((((......))))))))(((((((((.(......).)))))).))).))))..)........... ( -33.10) >DroMoj_CAF1 16593 120 - 1 AGCUGGUGCGCAACACCAGCCUGGAGGCCACCACUAUUCUGGCGCCCAUUUUGAUCAACUGCCAUUUAUCAGUGGCCAAUGCGCCCAAUGCAAUUGGCCCAUUAGGACCAUAUUUUCCAC .(((((((.....))))))).((((((((((........(((((...............))))).......))))))..((((.....))))..(((.((....)).))).....)))). ( -37.12) >DroAna_CAF1 13349 120 - 1 AGCUGGUGCGCAACACCAGUCUAGAAGCCACCAGCAUUUUGGCACCCAUUCUGAUCAAUUGUCAUCUUUCUGUCGCGAACGCUCCCAAUGCCAUUGGUCCCUUGGGACCCUAUUUUCCAC .(((((((.((..(.........)..)))))))))....(((((.......((((.....))))..........((....))......)))))..(((((....)))))........... ( -32.60) >consensus AGCUGGUGCGCAACACCAGCCUGGAAGCCACCAGCAUUCUGGCGCCCAUUCUGAUCAACUGCCAUUUGUCAGUGGCCAAUGCACCCAAUGCAAUUGGCCCAUUGGGACCAUAUUUUCCAC .(((((((.....))))))).((((((((((..(((...(((((...............)))))..)))..)))))....((.......))...(((.((....)).)))....))))). (-24.22 = -24.53 + 0.31)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:50:55 2006