
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 20,443,318 – 20,443,432 |
| Length | 114 |
| Max. P | 0.995166 |

| Location | 20,443,318 – 20,443,416 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 81.82 |
| Mean single sequence MFE | -33.68 |
| Consensus MFE | -27.16 |
| Energy contribution | -27.22 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.15 |
| Mean z-score | -3.59 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.963493 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20443318 98 + 27905053 CCCU--G----CGUCCCGAUGACGUGAACAACAACGAAAACGAUGAAAAUUUUUGCCAGAGUGCUACAGCAGAUAGCUUCAAUUCUGCUGUGGCACUCCGAACC ....--(----((((.....)))))......((.((....)).)).............(((((((((((((((..........)))))))))))))))...... ( -32.90) >DroVir_CAF1 31436 95 + 1 ---C--A----GAGGCAGACGAUGUGAAUAACAAUGAGAAUAGUGAGAAUUUUUGCCAGAGUGCUACAGCAGAUAGCUUCAAUGCUGCUGUGGCACUCACAACC ---.--.----..((((((((.(((.....))).)).....(((....))))))))).((((((((((((((.((.......))))))))))))))))...... ( -33.30) >DroGri_CAF1 24472 95 + 1 ---U--G----GAAGCGGACGAUGUCAAUAACAAUGAGAAUGGCGAGAAUUUUUGCCAGAGUGCUACUGCAGAUAGCUUCAAUGCUGCUGUAGCACUCGCAACC ---.--(----(..((.(((...)))..............(((((((....)))))))(((((((((.((((.((.......)))))).)))))))))))..)) ( -33.00) >DroYak_CAF1 26846 98 + 1 CCCA--G----CGUCCCGAUGACGUCAAUAACAACGAAAACGAUGAAAAUUUUUGCCAGAGUGCUACAGCAGAUAGCUUCAAUGCUGCUGUGGCACUCCGAACC ....--(----((((.....)))))......((.((....)).)).............((((((((((((((.((.......))))))))))))))))...... ( -30.30) >DroMoj_CAF1 24604 95 + 1 ---C--G----GAAGCGGACGAUGUGAACAACAAUGAGAAUGGCGAGAAUUUUUGCCAGAGUGCUACAGCAGAUAGCUUCAAUUCUGCUGUGGCACUCACAACC ---(--(----....))..((.(((.....))).))....(((((((....)))))))(((((((((((((((..........)))))))))))))))...... ( -38.00) >DroAna_CAF1 25599 104 + 1 CCACCACCCCCAAGUCCGAUGAUGUCAACAACAAUGCGAACGGUGAAAAUUUUUGCCAGAGUGCUACAGCAGAUAGCUUCAAUUCUGCUGUGGCACUCCGAACU .............((.((.((.((....)).))...)).))((..((....))..)).(((((((((((((((..........)))))))))))))))...... ( -34.60) >consensus ___C__G____GAAGCCGACGAUGUCAACAACAAUGAGAACGGUGAAAAUUUUUGCCAGAGUGCUACAGCAGAUAGCUUCAAUGCUGCUGUGGCACUCCCAACC .........................................((((((....)))))).((((((((((((((............))))))))))))))...... (-27.16 = -27.22 + 0.06)

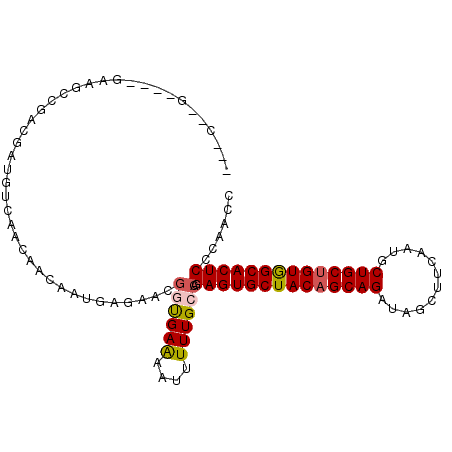

| Location | 20,443,318 – 20,443,416 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 81.82 |
| Mean single sequence MFE | -31.60 |
| Consensus MFE | -20.30 |
| Energy contribution | -20.47 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.790145 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20443318 98 - 27905053 GGUUCGGAGUGCCACAGCAGAAUUGAAGCUAUCUGCUGUAGCACUCUGGCAAAAAUUUUCAUCGUUUUCGUUGUUGUUCACGUCAUCGGGACG----C--AGGG .((.((((((((.((((((((..........)))))))).))))))))))..................(.((((.((((.((....)))))))----)--)).) ( -33.40) >DroVir_CAF1 31436 95 - 1 GGUUGUGAGUGCCACAGCAGCAUUGAAGCUAUCUGCUGUAGCACUCUGGCAAAAAUUCUCACUAUUCUCAUUGUUAUUCACAUCGUCUGCCUC----U--G--- ......((((((.(((((((.((.......))))))))).)))))).((((....................................))))..----.--.--- ( -23.52) >DroGri_CAF1 24472 95 - 1 GGUUGCGAGUGCUACAGCAGCAUUGAAGCUAUCUGCAGUAGCACUCUGGCAAAAAUUCUCGCCAUUCUCAUUGUUAUUGACAUCGUCCGCUUC----C--A--- ((..(((((((((((.((((.((.......)))))).)))))))))((((..........))))........................))..)----)--.--- ( -29.60) >DroYak_CAF1 26846 98 - 1 GGUUCGGAGUGCCACAGCAGCAUUGAAGCUAUCUGCUGUAGCACUCUGGCAAAAAUUUUCAUCGUUUUCGUUGUUAUUGACGUCAUCGGGACG----C--UGGG .((.((((((((.(((((((.((.......))))))))).))))))))))........(((.((((((((((......)))).....))))))----.--))). ( -31.70) >DroMoj_CAF1 24604 95 - 1 GGUUGUGAGUGCCACAGCAGAAUUGAAGCUAUCUGCUGUAGCACUCUGGCAAAAAUUCUCGCCAUUCUCAUUGUUGUUCACAUCGUCCGCUUC----C--G--- ((.(((((((((.((((((((..........)))))))).))))..((((..........)))).............)))))....)).....----.--.--- ( -29.30) >DroAna_CAF1 25599 104 - 1 AGUUCGGAGUGCCACAGCAGAAUUGAAGCUAUCUGCUGUAGCACUCUGGCAAAAAUUUUCACCGUUCGCAUUGUUGUUGACAUCAUCGGACUUGGGGGUGGUGG .((.((((((((.((((((((..........)))))))).)))))))))).........(((((..(.((...(((.((....)).)))...)).)..))))). ( -42.10) >consensus GGUUCGGAGUGCCACAGCAGAAUUGAAGCUAUCUGCUGUAGCACUCUGGCAAAAAUUCUCACCAUUCUCAUUGUUAUUCACAUCAUCGGCAUC____C__G___ .((...((((((.(((((((............))))))).))))))..))...................................................... (-20.30 = -20.47 + 0.17)



| Location | 20,443,336 – 20,443,432 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 85.95 |
| Mean single sequence MFE | -32.89 |
| Consensus MFE | -28.65 |
| Energy contribution | -29.23 |
| Covariance contribution | 0.59 |
| Combinations/Pair | 1.12 |
| Mean z-score | -4.67 |
| Structure conservation index | 0.87 |
| SVM decision value | 2.55 |
| SVM RNA-class probability | 0.995166 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20443336 96 + 27905053 UGAACAACAACGAAAACGAUGAAAAUUUUUGCCAGAGUGCUACAGCAGAUAGCUUCAAUUCUGCUGUGGCACUCCGAACCAAUAAAAUUGAAGCAA-------- .......((.((....)).)).......((((..(((((((((((((((..........))))))))))))))).....((((...))))..))))-------- ( -29.60) >DroVir_CAF1 31451 96 + 1 UGAAUAACAAUGAGAAUAGUGAGAAUUUUUGCCAGAGUGCUACAGCAGAUAGCUUCAAUGCUGCUGUGGCACUCACAACCAAAAAAAUUGAAGCAG-------- .......((((((((((.......))))))....((((((((((((((.((.......))))))))))))))))............))))......-------- ( -27.90) >DroPse_CAF1 35679 104 + 1 UCAACAACAAUGCCAACGAUGAGAAUUUUUGCCAGAGUGCUACAGCAGAUAGCUUCAAUUCUGCUGUGGCACUCCAAACAAACGAAAUUGAAGCAAACGCAGGC ...........(((..((((.......((((...(((((((((((((((..........)))))))))))))))))))........))))..((....)).))) ( -33.86) >DroGri_CAF1 24487 96 + 1 UCAAUAACAAUGAGAAUGGCGAGAAUUUUUGCCAGAGUGCUACUGCAGAUAGCUUCAAUGCUGCUGUAGCACUCGCAACCAAACAAAUUGAAGCAG-------- (((((...........(((((((....)))))))(((((((((.((((.((.......)))))).)))))))))............))))).....-------- ( -31.90) >DroMoj_CAF1 24619 96 + 1 UGAACAACAAUGAGAAUGGCGAGAAUUUUUGCCAGAGUGCUACAGCAGAUAGCUUCAAUUCUGCUGUGGCACUCACAACCAACCAAAUUGAAGCAG-------- .......((((.....(((((((....)))))))(((((((((((((((..........)))))))))))))))............))))......-------- ( -37.20) >DroAna_CAF1 25623 96 + 1 UCAACAACAAUGCGAACGGUGAAAAUUUUUGCCAGAGUGCUACAGCAGAUAGCUUCAAUUCUGCUGUGGCACUCCGAACUGACGAUAUUGAAGCAA-------- .......(((((((...((..((....))..)).(((((((((((((((..........)))))))))))))))........)).)))))......-------- ( -36.90) >consensus UCAACAACAAUGAGAACGGUGAGAAUUUUUGCCAGAGUGCUACAGCAGAUAGCUUCAAUUCUGCUGUGGCACUCCCAACCAACAAAAUUGAAGCAA________ .......((((......((((((....)))))).(((((((((((((((..........)))))))))))))))............)))).............. (-28.65 = -29.23 + 0.59)

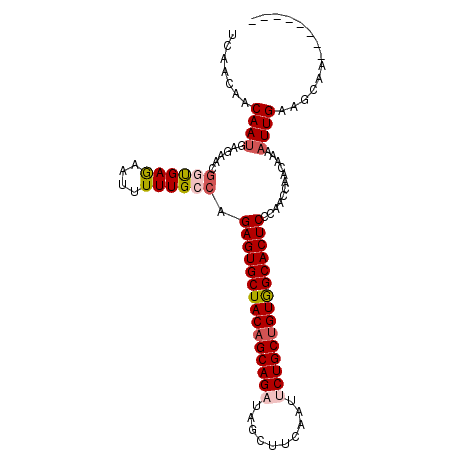

| Location | 20,443,336 – 20,443,432 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 85.95 |
| Mean single sequence MFE | -29.27 |
| Consensus MFE | -21.77 |
| Energy contribution | -22.77 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.86 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.710968 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20443336 96 - 27905053 --------UUGCUUCAAUUUUAUUGGUUCGGAGUGCCACAGCAGAAUUGAAGCUAUCUGCUGUAGCACUCUGGCAAAAAUUUUCAUCGUUUUCGUUGUUGUUCA --------((((.(((((...)))))..((((((((.((((((((..........)))))))).)))))))))))).......((.((....)).))....... ( -29.30) >DroVir_CAF1 31451 96 - 1 --------CUGCUUCAAUUUUUUUGGUUGUGAGUGCCACAGCAGCAUUGAAGCUAUCUGCUGUAGCACUCUGGCAAAAAUUCUCACUAUUCUCAUUGUUAUUCA --------.((((.(((((.....))))).((((((.(((((((.((.......))))))))).)))))).))))............................. ( -24.90) >DroPse_CAF1 35679 104 - 1 GCCUGCGUUUGCUUCAAUUUCGUUUGUUUGGAGUGCCACAGCAGAAUUGAAGCUAUCUGCUGUAGCACUCUGGCAAAAAUUCUCAUCGUUGGCAUUGUUGUUGA (((.(((((((((.(((......)))...(((((((.((((((((..........)))))))).))))))))))))).........))).)))........... ( -34.70) >DroGri_CAF1 24487 96 - 1 --------CUGCUUCAAUUUGUUUGGUUGCGAGUGCUACAGCAGCAUUGAAGCUAUCUGCAGUAGCACUCUGGCAAAAAUUCUCGCCAUUCUCAUUGUUAUUGA --------..((.((((.....))))..))(((((((((.((((.((.......)))))).)))))))))((((..........))))................ ( -27.50) >DroMoj_CAF1 24619 96 - 1 --------CUGCUUCAAUUUGGUUGGUUGUGAGUGCCACAGCAGAAUUGAAGCUAUCUGCUGUAGCACUCUGGCAAAAAUUCUCGCCAUUCUCAUUGUUGUUCA --------..((..((((..(..(((((((((((((.((((((((..........)))))))).))))))..))).........))))..)..))))..))... ( -29.20) >DroAna_CAF1 25623 96 - 1 --------UUGCUUCAAUAUCGUCAGUUCGGAGUGCCACAGCAGAAUUGAAGCUAUCUGCUGUAGCACUCUGGCAAAAAUUUUCACCGUUCGCAUUGUUGUUGA --------.....((((((....((((.((((((((.((((((((..........)))))))).))))))))((.................)))))).)))))) ( -30.03) >consensus ________CUGCUUCAAUUUCGUUGGUUGGGAGUGCCACAGCAGAAUUGAAGCUAUCUGCUGUAGCACUCUGGCAAAAAUUCUCACCAUUCUCAUUGUUGUUCA .........((((((((.....))))....((((((.((((((((..........)))))))).)))))).))))............................. (-21.77 = -22.77 + 1.00)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:50:44 2006