
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 20,437,424 – 20,437,517 |
| Length | 93 |
| Max. P | 0.758243 |

| Location | 20,437,424 – 20,437,517 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 85.84 |
| Mean single sequence MFE | -25.45 |
| Consensus MFE | -20.20 |
| Energy contribution | -20.32 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.736159 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
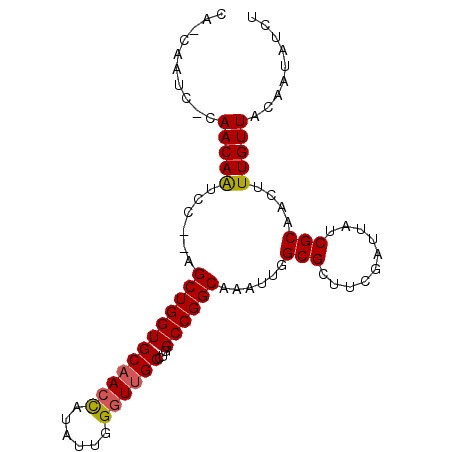
>3R_DroMel_CAF1 20437424 93 + 27905053 CA-CAAUCCAAACAAUUCCUGGCUGGUGCAACUAUAUUGGGUUGCCUGGCCGGCAAAUUGGCGUUUCGAUUAUCGCAACUUUGUUACAAUAUCU .(-(((......(((((.(((((..(.((((((......)))))))..)))))..)))))(((..........)))....)))).......... ( -26.90) >DroSim_CAF1 19590 85 + 1 ---------GAACAAUCCCUGGCUGGUGCAACCAUAUUGGGUUGCCUGGCCGGCAAAUUGGCGUUUCGAUUAUCGCAACUUUGUUACAAUAUCU ---------.(((((...(((((..(.((((((......)))))))..))))).......(((..........)))....)))))......... ( -27.40) >DroEre_CAF1 19528 90 + 1 CAGCAAUC-CAACAAUCC--AGCUGGUGCAACCAUAUUGGUUGGCCUGGCCGGCA-AUCGGCGCUUCGAUUAUCGCAACUUUGUUACGUUAUCU .(((....-...((..((--(((..(((......)))..)))))..))((((...-..)))))))..(((...((.(((...))).))..))). ( -22.50) >DroYak_CAF1 20676 91 + 1 GAGCAAUC-CAACAGUUC--AGCUGGUGCAACCAUAUUGGGUUGCCUGGCCGGCAAAUUGGCGCUUCGAUUAUCGCAACUUUGUUACAAUAUCU .(((((..-...(((((.--.((((((((((((......))))))...)))))).)))))(((.(......).)))....)))))......... ( -25.00) >consensus CA_CAAUC_CAACAAUCC__AGCUGGUGCAACCAUAUUGGGUUGCCUGGCCGGCAAAUUGGCGCUUCGAUUAUCGCAACUUUGUUACAAUAUCU ..........(((((......((((((((((((......))))))...))))))......(((..........)))....)))))......... (-20.20 = -20.32 + 0.12)


| Location | 20,437,424 – 20,437,517 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 85.84 |
| Mean single sequence MFE | -27.08 |
| Consensus MFE | -18.48 |
| Energy contribution | -19.22 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.758243 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20437424 93 - 27905053 AGAUAUUGUAACAAAGUUGCGAUAAUCGAAACGCCAAUUUGCCGGCCAGGCAACCCAAUAUAGUUGCACCAGCCAGGAAUUGUUUGGAUUG-UG .(((((((((((...)))))))).)))......((((....(((((..((((((........))))).)..))).))......))))....-.. ( -23.80) >DroSim_CAF1 19590 85 - 1 AGAUAUUGUAACAAAGUUGCGAUAAUCGAAACGCCAAUUUGCCGGCCAGGCAACCCAAUAUGGUUGCACCAGCCAGGGAUUGUUC--------- .(((((((((((...)))))))).))).......(((((..(((((..(((((((......)))))).)..))).)))))))...--------- ( -24.90) >DroEre_CAF1 19528 90 - 1 AGAUAACGUAACAAAGUUGCGAUAAUCGAAGCGCCGAU-UGCCGGCCAGGCCAACCAAUAUGGUUGCACCAGCU--GGAUUGUUG-GAUUGCUG .(((..((((((...))))))...)))..(((.(((((-..(((((..((.(((((.....)))))..)).)))--))...))))-)...))). ( -30.20) >DroYak_CAF1 20676 91 - 1 AGAUAUUGUAACAAAGUUGCGAUAAUCGAAGCGCCAAUUUGCCGGCCAGGCAACCCAAUAUGGUUGCACCAGCU--GAACUGUUG-GAUUGCUC .(((((((((((...)))))))).)))..(((.(((((....((((..(((((((......)))))).)..)))--)....))))-)...))). ( -29.40) >consensus AGAUAUUGUAACAAAGUUGCGAUAAUCGAAACGCCAAUUUGCCGGCCAGGCAACCCAAUAUGGUUGCACCAGCC__GAAUUGUUG_GAUUG_UG .(((((((((((...)))))))).))).......((((((...(((..(((((((......)))))).)..)))..))))))............ (-18.48 = -19.22 + 0.75)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:50:38 2006