
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 20,435,901 – 20,436,032 |
| Length | 131 |
| Max. P | 0.987662 |

| Location | 20,435,901 – 20,436,008 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 84.10 |
| Mean single sequence MFE | -26.73 |
| Consensus MFE | -22.65 |
| Energy contribution | -22.77 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.927618 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20435901 107 + 27905053 ACUGGGUAUUCCCCGGACCGGGCAAAAUGCUAACCAUUUACCGCCCAAAACGACGCC----CCCUGGUUACACCCCCACUCCAAACAUUCCCCCCACUUCCGUGGUGGGCU .(((((.....)))))...((((.(((((.....)))))...))))........(((----(..(((........)))...............((((....)))).)))). ( -28.70) >DroSec_CAF1 18391 105 + 1 ACUGGGUAUUCCCCGGACCGGGCAAAAUGCUAACCAUUUACCGCCCAAAACGCCGCC----CCCUGGUUACAACCCCACUCCAAGCAU--CCCCCACUUCCAUGGUGGGCU .(((((.....)))))...((((.(((((.....)))))...))))........(((----((((((.....................--.........))).)).)))). ( -27.61) >DroSim_CAF1 18045 105 + 1 ACUGGGUAUUCCCCGGACCGGGCAAAAUGCUAACCAUUUACCGCCCAAAACGCCGCC----CCCUGGUUACAACCCCACUCCAGCCAC--CCCCCACUUCCAUGGUGGGCU .(((((.....)))))...((((.(((((.....)))))...))))...........----..((((.............))))....--..((((((.....)))))).. ( -27.62) >DroYak_CAF1 19086 108 + 1 CCUGGGUAUUCCCCGGACCGGGCAAAAUGCUAACCAUUUACCGCCCAAAACGGAACCCACACCCAAACCUCCACCCC-CUUCAACAGC--CUUCCCCCUUCUUGCUGGGCU ..(((((.....(((....((((.(((((.....)))))...))))....))).......)))))............-.......(((--((..(........)..))))) ( -23.00) >consensus ACUGGGUAUUCCCCGGACCGGGCAAAAUGCUAACCAUUUACCGCCCAAAACGCCGCC____CCCUGGUUACAACCCCACUCCAACCAC__CCCCCACUUCCAUGGUGGGCU .(((((.....)))))...((((.(((((.....)))))...))))..............................................((((((.....)))))).. (-22.65 = -22.77 + 0.13)


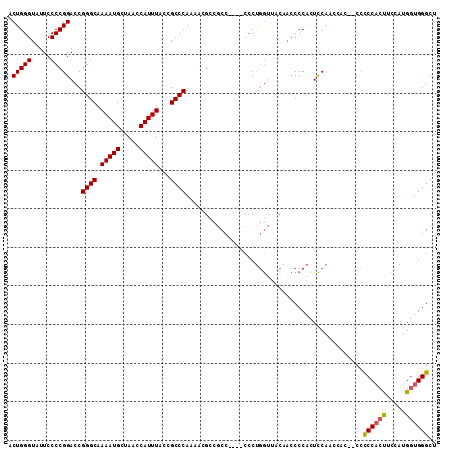
| Location | 20,435,901 – 20,436,008 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 84.10 |
| Mean single sequence MFE | -41.38 |
| Consensus MFE | -25.29 |
| Energy contribution | -25.60 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.701507 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20435901 107 - 27905053 AGCCCACCACGGAAGUGGGGGGAAUGUUUGGAGUGGGGGUGUAACCAGGG----GGCGUCGUUUUGGGCGGUAAAUGGUUAGCAUUUUGCCCGGUCCGGGGAAUACCCAGU ..(((.((((....)))))))........(((.((((((((((((((...----(.((((......)))).)...))))).)))))...)))).)))(((.....)))... ( -46.90) >DroSec_CAF1 18391 105 - 1 AGCCCACCAUGGAAGUGGGGG--AUGCUUGGAGUGGGGUUGUAACCAGGG----GGCGGCGUUUUGGGCGGUAAAUGGUUAGCAUUUUGCCCGGUCCGGGGAAUACCCAGU ..(((.((((....)))))))--......(((.((((..((((((((...----.((.((.......)).))...))))).))).....)))).)))(((.....)))... ( -38.30) >DroSim_CAF1 18045 105 - 1 AGCCCACCAUGGAAGUGGGGG--GUGGCUGGAGUGGGGUUGUAACCAGGG----GGCGGCGUUUUGGGCGGUAAAUGGUUAGCAUUUUGCCCGGUCCGGGGAAUACCCAGU .((((.((((....)))).))--))((((((.(..((((..((((((...----.((.((.......)).))...))))))..))))..))))))).(((.....)))... ( -42.20) >DroYak_CAF1 19086 108 - 1 AGCCCAGCAAGAAGGGGGAAG--GCUGUUGAAG-GGGGUGGAGGUUUGGGUGUGGGUUCCGUUUUGGGCGGUAAAUGGUUAGCAUUUUGCCCGGUCCGGGGAAUACCCAGG .((((..(((..((.......--.)).)))...-.))))......((((((((...(((((..(((((((..(((((.....))))))))))))..))))).)))))))). ( -38.10) >consensus AGCCCACCAUGGAAGUGGGGG__AUGGUUGGAGUGGGGUUGUAACCAGGG____GGCGGCGUUUUGGGCGGUAAAUGGUUAGCAUUUUGCCCGGUCCGGGGAAUACCCAGU .(((..((((....))))...............(((........))).......)))..((..(((((((..(((((.....))))))))))))..))(((....)))... (-25.29 = -25.60 + 0.31)



| Location | 20,435,941 – 20,436,032 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 80.04 |
| Mean single sequence MFE | -26.85 |
| Consensus MFE | -20.15 |
| Energy contribution | -21.14 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.75 |
| SVM decision value | 2.09 |
| SVM RNA-class probability | 0.987662 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20435941 91 + 27905053 CCGCCCAAAACGACGCC----CCCUGGUUACACCCCCACUCCAAACAUUCCCCCCACUUCCGUGGUGGGCU---GCUGUGUUGUGGGCUUGUCUAAUU ..((((...(((((((.----...(((........)))............(((((((....)))).)))..---...))))))))))).......... ( -26.10) >DroSec_CAF1 18431 89 + 1 CCGCCCAAAACGCCGCC----CCCUGGUUACAACCCCACUCCAAGCAU--CCCCCACUUCCAUGGUGGGCU---GCUGUGUUGUGGGCUUGUCUAAUU ..(((((.(((((....----....(((....)))........((((.--((((((......))).))).)---)))))))).))))).......... ( -29.30) >DroSim_CAF1 18085 89 + 1 CCGCCCAAAACGCCGCC----CCCUGGUUACAACCCCACUCCAGCCAC--CCCCCACUUCCAUGGUGGGCU---GCUGUGUUGUGGGCUUGUCUAAUU ..(((((.(((((.((.----..((((.............))))....--..((((((.....))))))..---)).))))).))))).......... ( -29.92) >DroYak_CAF1 19126 95 + 1 CCGCCCAAAACGGAACCCACACCCAAACCUCCACCCC-CUUCAACAGC--CUUCCCCCUUCUUGCUGGGCUGCUGCUAUGUUGUGGGCUUGUCUAAUU ..(((((.((((((...............))).....-...((.((((--((..(........)..)))))).))....))).))))).......... ( -22.06) >consensus CCGCCCAAAACGCCGCC____CCCUGGUUACAACCCCACUCCAACCAC__CCCCCACUUCCAUGGUGGGCU___GCUGUGUUGUGGGCUUGUCUAAUU ..(((((.(((((.((........(((.............))).........((((((.....)))))).....)).))))).))))).......... (-20.15 = -21.14 + 1.00)



| Location | 20,435,941 – 20,436,032 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 80.04 |
| Mean single sequence MFE | -34.65 |
| Consensus MFE | -21.86 |
| Energy contribution | -22.17 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.63 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.926802 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20435941 91 - 27905053 AAUUAGACAAGCCCACAACACAGC---AGCCCACCACGGAAGUGGGGGGAAUGUUUGGAGUGGGGGUGUAACCAGGG----GGCGUCGUUUUGGGCGG ......(((..(((((..((.(((---(.(((.((((....)))))))...))))))..)))))..)))..((((((----.(...).)))))).... ( -35.10) >DroSec_CAF1 18431 89 - 1 AAUUAGACAAGCCCACAACACAGC---AGCCCACCAUGGAAGUGGGGG--AUGCUUGGAGUGGGGUUGUAACCAGGG----GGCGGCGUUUUGGGCGG ......((((.(((((..((.(((---(.(((.((((....)))))))--.))))))..))))).))))..((((((----.(...).)))))).... ( -35.90) >DroSim_CAF1 18085 89 - 1 AAUUAGACAAGCCCACAACACAGC---AGCCCACCAUGGAAGUGGGGG--GUGGCUGGAGUGGGGUUGUAACCAGGG----GGCGGCGUUUUGGGCGG ......((((.(((((....((((---.((((.((((....)))).))--)).))))..))))).))))..((((((----.(...).)))))).... ( -37.10) >DroYak_CAF1 19126 95 - 1 AAUUAGACAAGCCCACAACAUAGCAGCAGCCCAGCAAGAAGGGGGAAG--GCUGUUGAAG-GGGGUGGAGGUUUGGGUGUGGGUUCCGUUUUGGGCGG ..........(((((.(((.(((((((..(((..(.....).)))...--)))))))...-.....((((.((.......)).))))))).))))).. ( -30.50) >consensus AAUUAGACAAGCCCACAACACAGC___AGCCCACCAUGGAAGUGGGGG__AUGGUUGGAGUGGGGUUGUAACCAGGG____GGCGGCGUUUUGGGCGG ..........(((((.(((...((.....(((.((((....))))...............(((........)))))).....))...))).))))).. (-21.86 = -22.17 + 0.31)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:50:37 2006