
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 20,434,234 – 20,434,368 |
| Length | 134 |
| Max. P | 0.952286 |

| Location | 20,434,234 – 20,434,347 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 93.60 |
| Mean single sequence MFE | -14.46 |
| Consensus MFE | -12.44 |
| Energy contribution | -12.64 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.869565 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20434234 113 + 27905053 GCAAAAAAAAAAAAAGCGCAGCAUUUUAACAAGUUAAACAAUCAAAGAAA---UCGUAUUCAAGUGCCACAAAGUUGAAGUGGCGUAUAAAAUAAACAAAUAAUGCGAAUAUAAAU ................((((..((((.......(((.((.......(((.---.....)))....(((((.........))))))).))).......))))..))))......... ( -15.44) >DroSec_CAF1 16838 105 + 1 GUAAAA-------AAGAGCAGCAUUUUAACAAGU-AAACAAUCAAAGAAA---UCGUAUUCAAGUGCCACAAAGUUGAAGUGGCGUAUAAAAUAAACAAAUAAUGCGAAUAUAAAU ......-------.....................-...............---..((((((....(((((.........)))))((((..............)))))))))).... ( -13.04) >DroSim_CAF1 16499 105 + 1 GCAAAA-------AAGAGCAGCAUUUUAACAAGU-AAACAAUCAAAGAAA---UCGUAUUCAAGUGCCACAAAGUUGAAGUGGCGUAUAAAAUAAACAAAUAAUGCGAAUAUAAAU ((....-------....))...............-...............---..((((((....(((((.........)))))((((..............)))))))))).... ( -14.04) >DroEre_CAF1 16450 105 + 1 GCAAAA-------AAGAGCAACGUUUUAACAAGU-AAACAAUCAAAGAAA---UCGUAUUCAAGUGCCACAAAGUUGAAGUGGCGUAUAAAAUAAACAAAUAAUGCGAAUAUAAAU ((....-------....))...((((........-))))...........---..((((((....(((((.........)))))((((..............)))))))))).... ( -15.24) >DroYak_CAF1 17474 108 + 1 GCAAAA-------AAGAGCAACAUUUUAACAAGU-AAACAAUCAAAGAUAUCAUCGUACUCAAGUGCCACAAAGUUGAAGUGGCGUAUAAAAUAAACAAAUAAUGCGAAUAUAAAU ((....-------....))..(((((((((....-...........(((...)))((((....))))......))))))))).(((((..............)))))......... ( -14.54) >consensus GCAAAA_______AAGAGCAGCAUUUUAACAAGU_AAACAAUCAAAGAAA___UCGUAUUCAAGUGCCACAAAGUUGAAGUGGCGUAUAAAAUAAACAAAUAAUGCGAAUAUAAAU .......................................................((((((....(((((.........)))))((((..............)))))))))).... (-12.44 = -12.64 + 0.20)



| Location | 20,434,273 – 20,434,368 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 75.53 |
| Mean single sequence MFE | -17.71 |
| Consensus MFE | -10.83 |
| Energy contribution | -10.97 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.952286 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20434273 95 + 27905053 AAUCAAAGAAA---UCGUAUUCAAGUGCCACAAAG-UUGAAGUGGCGUAUAAAAUAAACAAAUAAUGCGAAUAUAAAUCAAGAUACGCA-------------------ACUUGAAAAG ...........---.....((((((((((((....-.....)))))...................((((.((..........)).))))-------------------)))))))... ( -17.50) >DroSec_CAF1 16869 95 + 1 AAUCAAAGAAA---UCGUAUUCAAGUGCCACAAAG-UUGAAGUGGCGUAUAAAAUAAACAAAUAAUGCGAAUAUAAAUAAAGAUAUGCU-------------------ACUUGAAAAG ...........---.....((((((((((((....-.....)))))((((..............)))).....................-------------------)))))))... ( -15.04) >DroSim_CAF1 16530 95 + 1 AAUCAAAGAAA---UCGUAUUCAAGUGCCACAAAG-UUGAAGUGGCGUAUAAAAUAAACAAAUAAUGCGAAUAUAAAUAAAGAUAUGCA-------------------ACUUGAAAAG ...........---.....((((((((((((....-.....)))))...................(((..((((........)))))))-------------------)))))))... ( -16.20) >DroEre_CAF1 16481 108 + 1 AAUCAAAGAAA---UCGUAUUCAAGUGCCACAAAG-UUGAAGUGGCGUAUAAAAUAAACAAAUAAUGCGAAUAUAAAUAUAGAUAUGCA------ACUCACAGCGACGACUUGAAAAG ...........---.....((((((((((((....-.....)))))...................((((..(((....)))....))))------.............)))))))... ( -16.50) >DroYak_CAF1 17505 114 + 1 AAUCAAAGAUAUCAUCGUACUCAAGUGCCACAAAG-UUGAAGUGGCGUAUAAAAUAAACAAAUAAUGCGAAUAUAAAUAUAGAUAUGCA--G-CGAUUCACAGCGACGUCUUGAAAAG ..((((.((..((.(((((.......(((((....-.....)))))((.........))......)))))...................--(-(........))))..)))))).... ( -19.00) >DroAna_CAF1 17444 112 + 1 AGUAAUACAGA---UCUUUUUCAAGUGCUGCCUACAUU---GUGUUGAAAGAAGCGAAAAAAUAUUACGAAAAUAAAGUGAGAUAUCAGAAGUCGGCAUGCAUGAAAGAGUUGAAAUG .......(((.---.((((((((..(((((((.((.((---((.((....)).)))).......((((.........))))..........)).)))).))))))))))))))..... ( -22.00) >consensus AAUCAAAGAAA___UCGUAUUCAAGUGCCACAAAG_UUGAAGUGGCGUAUAAAAUAAACAAAUAAUGCGAAUAUAAAUAAAGAUAUGCA___________________ACUUGAAAAG ...................((((((((((((..........)))))((((..............))))........................................)))))))... (-10.83 = -10.97 + 0.14)

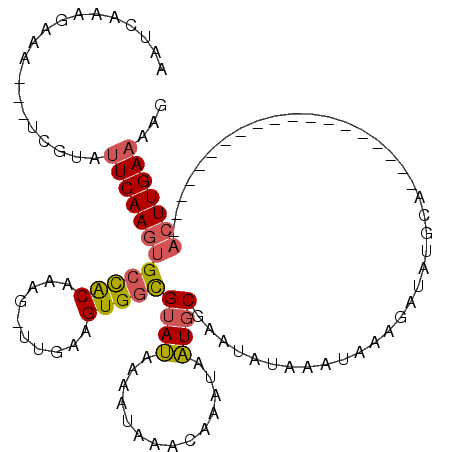

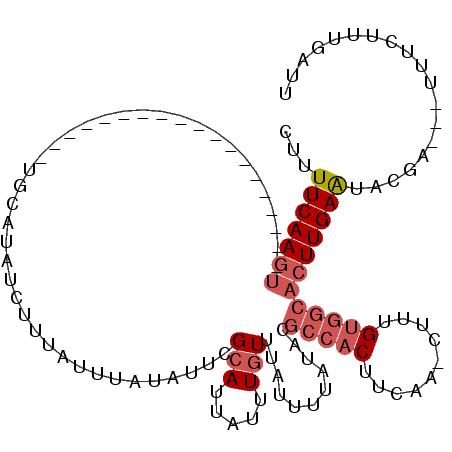
| Location | 20,434,273 – 20,434,368 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 75.53 |
| Mean single sequence MFE | -16.94 |
| Consensus MFE | -8.97 |
| Energy contribution | -10.00 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.705769 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20434273 95 - 27905053 CUUUUCAAGU-------------------UGCGUAUCUUGAUUUAUAUUCGCAUUAUUUGUUUAUUUUAUACGCCACUUCAA-CUUUGUGGCACUUGAAUACGA---UUUCUUUGAUU ...(((((((-------------------((((................))))...................(((((.....-....)))))))))))).....---........... ( -17.49) >DroSec_CAF1 16869 95 - 1 CUUUUCAAGU-------------------AGCAUAUCUUUAUUUAUAUUCGCAUUAUUUGUUUAUUUUAUACGCCACUUCAA-CUUUGUGGCACUUGAAUACGA---UUUCUUUGAUU ...(((((((-------------------.....................(((.....)))...........(((((.....-....)))))))))))).....---........... ( -15.40) >DroSim_CAF1 16530 95 - 1 CUUUUCAAGU-------------------UGCAUAUCUUUAUUUAUAUUCGCAUUAUUUGUUUAUUUUAUACGCCACUUCAA-CUUUGUGGCACUUGAAUACGA---UUUCUUUGAUU ...(((((((-------------------(((((((........))))..)))...................(((((.....-....)))))))))))).....---........... ( -15.20) >DroEre_CAF1 16481 108 - 1 CUUUUCAAGUCGUCGCUGUGAGU------UGCAUAUCUAUAUUUAUAUUCGCAUUAUUUGUUUAUUUUAUACGCCACUUCAA-CUUUGUGGCACUUGAAUACGA---UUUCUUUGAUU ...(((((((.(((((...((((------((......((((...(((...(((.....))).)))..))))........)))-))).)))))))))))).....---........... ( -19.64) >DroYak_CAF1 17505 114 - 1 CUUUUCAAGACGUCGCUGUGAAUCG-C--UGCAUAUCUAUAUUUAUAUUCGCAUUAUUUGUUUAUUUUAUACGCCACUUCAA-CUUUGUGGCACUUGAGUACGAUGAUAUCUUUGAUU ....((((((..((((.(((...))-)--.))..........(..((((((((.....)))...........(((((.....-....)))))....)))))..).))..)).)))).. ( -21.00) >DroAna_CAF1 17444 112 - 1 CAUUUCAACUCUUUCAUGCAUGCCGACUUCUGAUAUCUCACUUUAUUUUCGUAAUAUUUUUUCGCUUCUUUCAACAC---AAUGUAGGCAGCACUUGAAAAAGA---UCUGUAUUACU ........((.((((((((.((((.((...(((....)))...((((.....)))).....................---...)).)))))))..))))).)).---........... ( -12.90) >consensus CUUUUCAAGU___________________UGCAUAUCUUUAUUUAUAUUCGCAUUAUUUGUUUAUUUUAUACGCCACUUCAA_CUUUGUGGCACUUGAAUACGA___UUUCUUUGAUU ...(((((((........................................(((.....)))...........(((((..........))))))))))))................... ( -8.97 = -10.00 + 1.03)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:50:32 2006