
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 20,426,429 – 20,426,565 |
| Length | 136 |
| Max. P | 0.999990 |

| Location | 20,426,429 – 20,426,525 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 89.21 |
| Mean single sequence MFE | -27.32 |
| Consensus MFE | -25.40 |
| Energy contribution | -25.56 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.93 |
| SVM decision value | 3.87 |
| SVM RNA-class probability | 0.999674 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20426429 96 + 27905053 UACAUACAUAUAGAAGCCGAA-------AUCGGCGGUGCAAGAUAAAGCGGGCUAAUUACAUAACGUUAGCACCGCGUACUGGUUAAGUGUAGAAACCACUGC ....(((((.(((..((((..-------..))))(((((........((((((((((........)))))).)))))))))..))).)))))........... ( -27.00) >DroSec_CAF1 9437 93 + 1 ---AUACAUAUAGAAGCCAAA-------AUCGGCGGUGCAAGAUAAAGCGGGCUAAUUACAUAACGUUAGCACCGCGUACUGGUUAAGUGUAGAAACCACUGC ---.(((((.(((..(((...-------...)))(((((........((((((((((........)))))).)))))))))..))).)))))........... ( -24.20) >DroSim_CAF1 9024 93 + 1 ---AUACAUAUAGAAGCCGAA-------AUCGGCGGUGCAAGAUAAAGCGGGCUAAUUACAUAACGUUAGCACCGCGUACUGGUUAAGUGUAGAAACCACUGC ---.(((((.(((..((((..-------..))))(((((........((((((((((........)))))).)))))))))..))).)))))........... ( -27.00) >DroEre_CAF1 8930 87 + 1 ---------UUGGAAGCCGAA-------AUCGGCGGUGCAAGAUAAAGCGGGCUAAUUACAUAACGUUAGCACCGCGCACUGGUUAAGUGUAGAAACCAUUGC ---------......((((..-------..))))(((..........((((((((((........)))))).))))(((((.....)))))....)))..... ( -30.20) >DroYak_CAF1 9458 94 + 1 ---------AUAGAAGCCGAAAGCCAAAAACGGCGGUGCAAGAUAAAGCGGGCUAAUUACAUAACGUUAGCACCGCGCACUGGUUAAGUGUAGAAACCAUUGC ---------......((((...........))))(((..........((((((((((........)))))).))))(((((.....)))))....)))..... ( -28.20) >consensus ___AUACAUAUAGAAGCCGAA_______AUCGGCGGUGCAAGAUAAAGCGGGCUAAUUACAUAACGUUAGCACCGCGUACUGGUUAAGUGUAGAAACCACUGC ...............(((((.........)))))(((..........((((((((((........)))))).))))(((((.....)))))....)))..... (-25.40 = -25.56 + 0.16)



| Location | 20,426,429 – 20,426,525 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 89.21 |
| Mean single sequence MFE | -25.16 |
| Consensus MFE | -25.14 |
| Energy contribution | -24.94 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.00 |
| Structure conservation index | 1.00 |
| SVM decision value | 3.18 |
| SVM RNA-class probability | 0.998678 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20426429 96 - 27905053 GCAGUGGUUUCUACACUUAACCAGUACGCGGUGCUAACGUUAUGUAAUUAGCCCGCUUUAUCUUGCACCGCCGAU-------UUCGGCUUCUAUAUGUAUGUA (((((((((.........)))))....((((.(((((..........)))))))))......))))...((((..-------..))))............... ( -25.30) >DroSec_CAF1 9437 93 - 1 GCAGUGGUUUCUACACUUAACCAGUACGCGGUGCUAACGUUAUGUAAUUAGCCCGCUUUAUCUUGCACCGCCGAU-------UUUGGCUUCUAUAUGUAU--- (((((((((.........)))))....((((.(((((..........)))))))))......))))...((((..-------..))))............--- ( -22.50) >DroSim_CAF1 9024 93 - 1 GCAGUGGUUUCUACACUUAACCAGUACGCGGUGCUAACGUUAUGUAAUUAGCCCGCUUUAUCUUGCACCGCCGAU-------UUCGGCUUCUAUAUGUAU--- (((((((((.........)))))....((((.(((((..........)))))))))......))))...((((..-------..))))............--- ( -25.30) >DroEre_CAF1 8930 87 - 1 GCAAUGGUUUCUACACUUAACCAGUGCGCGGUGCUAACGUUAUGUAAUUAGCCCGCUUUAUCUUGCACCGCCGAU-------UUCGGCUUCCAA--------- (((.(((((.........))))).)))((((.(((((..........))))))))).............((((..-------..))))......--------- ( -26.90) >DroYak_CAF1 9458 94 - 1 GCAAUGGUUUCUACACUUAACCAGUGCGCGGUGCUAACGUUAUGUAAUUAGCCCGCUUUAUCUUGCACCGCCGUUUUUGGCUUUCGGCUUCUAU--------- (((.(((((.........))))).)))((((.(((((..........)))))))))........((...((((....)))).....))......--------- ( -25.80) >consensus GCAGUGGUUUCUACACUUAACCAGUACGCGGUGCUAACGUUAUGUAAUUAGCCCGCUUUAUCUUGCACCGCCGAU_______UUCGGCUUCUAUAUGUAU___ (((((((((.........)))))....((((.(((((..........)))))))))......))))...(((((.........)))))............... (-25.14 = -24.94 + -0.20)

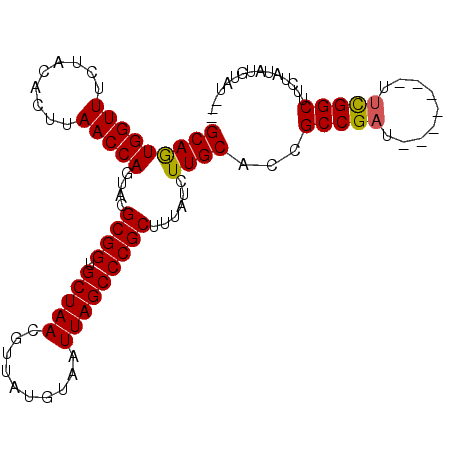

| Location | 20,426,462 – 20,426,565 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 93.91 |
| Mean single sequence MFE | -29.41 |
| Consensus MFE | -28.55 |
| Energy contribution | -28.15 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.95 |
| Structure conservation index | 0.97 |
| SVM decision value | 5.56 |
| SVM RNA-class probability | 0.999990 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20426462 103 + 27905053 AGAUAAAGCGGGCUAAUUACAUAACGUUAGCACCGCGUACUGGUUAAGUGUAGAAACCACUGCUUUUCGGCCAUUUCUAUACAGCUAUUAAAUUGGUCAAUUA .(((...((((((((((........)))))).))))....(((((..(((((((((...(((.....)))...))))))))))))))........)))..... ( -26.80) >DroSec_CAF1 9467 101 + 1 AGAUAAAGCGGGCUAAUUACAUAACGUUAGCACCGCGUACUGGUUAAGUGUAGAAACCACUGCUUUUCGGCCAUUUCU--ACUGCUAUUAACUUGGCCAAUUA .......((((((((((........)))))).))))....((((((((((((((((...(((.....)))...)))))--))........))))))))).... ( -29.40) >DroSim_CAF1 9054 101 + 1 AGAUAAAGCGGGCUAAUUACAUAACGUUAGCACCGCGUACUGGUUAAGUGUAGAAACCACUGCUUUUCGGCCAUUUCU--ACUGCUAUUAACUUGGCCAAUUA .......((((((((((........)))))).))))....((((((((((((((((...(((.....)))...)))))--))........))))))))).... ( -29.40) >DroEre_CAF1 8954 101 + 1 AGAUAAAGCGGGCUAAUUACAUAACGUUAGCACCGCGCACUGGUUAAGUGUAGAAACCAUUGCUUUUCGGCCAUUUCU--ACAGCUAUUUAAUUGGCCAAUUU .......((((((((((........)))))).))))(((.(((((.........))))).))).....(((((.....--.............)))))..... ( -30.07) >DroYak_CAF1 9489 101 + 1 AGAUAAAGCGGGCUAAUUACAUAACGUUAGCACCGCGCACUGGUUAAGUGUAGAAACCAUUGCUUUUUGGCCAUUUCU--ACGACCAUUUAAUUGGCCAAUUU .......((((((((((........)))))).))))(((.(((((.........))))).)))...(((((((.....--.............)))))))... ( -31.37) >consensus AGAUAAAGCGGGCUAAUUACAUAACGUUAGCACCGCGUACUGGUUAAGUGUAGAAACCACUGCUUUUCGGCCAUUUCU__ACAGCUAUUAAAUUGGCCAAUUA .......((((((((((........)))))).))))(((.(((((.........))))).))).....(((((....................)))))..... (-28.55 = -28.15 + -0.40)

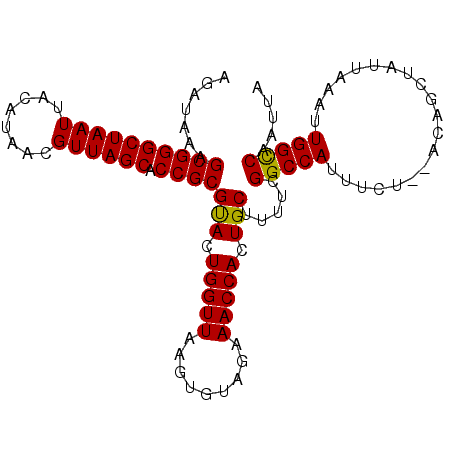

| Location | 20,426,462 – 20,426,565 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 93.91 |
| Mean single sequence MFE | -26.66 |
| Consensus MFE | -21.97 |
| Energy contribution | -22.01 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.931451 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20426462 103 - 27905053 UAAUUGACCAAUUUAAUAGCUGUAUAGAAAUGGCCGAAAAGCAGUGGUUUCUACACUUAACCAGUACGCGGUGCUAACGUUAUGUAAUUAGCCCGCUUUAUCU .....................((.(((((((.((.(.....).)).))))))).))...........((((.(((((..........)))))))))....... ( -20.30) >DroSec_CAF1 9467 101 - 1 UAAUUGGCCAAGUUAAUAGCAGU--AGAAAUGGCCGAAAAGCAGUGGUUUCUACACUUAACCAGUACGCGGUGCUAACGUUAUGUAAUUAGCCCGCUUUAUCU ..(((((..((((........((--((((((.((.(.....).)).))))))))))))..)))))..((((.(((((..........)))))))))....... ( -26.80) >DroSim_CAF1 9054 101 - 1 UAAUUGGCCAAGUUAAUAGCAGU--AGAAAUGGCCGAAAAGCAGUGGUUUCUACACUUAACCAGUACGCGGUGCUAACGUUAUGUAAUUAGCCCGCUUUAUCU ..(((((..((((........((--((((((.((.(.....).)).))))))))))))..)))))..((((.(((((..........)))))))))....... ( -26.80) >DroEre_CAF1 8954 101 - 1 AAAUUGGCCAAUUAAAUAGCUGU--AGAAAUGGCCGAAAAGCAAUGGUUUCUACACUUAACCAGUGCGCGGUGCUAACGUUAUGUAAUUAGCCCGCUUUAUCU ...(((((((.((..((....))--..)).)))))))...(((.(((((.........))))).)))((((.(((((..........)))))))))....... ( -29.20) >DroYak_CAF1 9489 101 - 1 AAAUUGGCCAAUUAAAUGGUCGU--AGAAAUGGCCAAAAAGCAAUGGUUUCUACACUUAACCAGUGCGCGGUGCUAACGUUAUGUAAUUAGCCCGCUUUAUCU ...(((((((.((..((....))--..)).)))))))...(((.(((((.........))))).)))((((.(((((..........)))))))))....... ( -30.20) >consensus UAAUUGGCCAAUUUAAUAGCAGU__AGAAAUGGCCGAAAAGCAGUGGUUUCUACACUUAACCAGUACGCGGUGCUAACGUUAUGUAAUUAGCCCGCUUUAUCU ...(((((((....................))))))).......(((((.........)))))....((((.(((((..........)))))))))....... (-21.97 = -22.01 + 0.04)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:50:28 2006