| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 2,669,434 – 2,669,534 |
| Length | 100 |
| Max. P | 0.588592 |

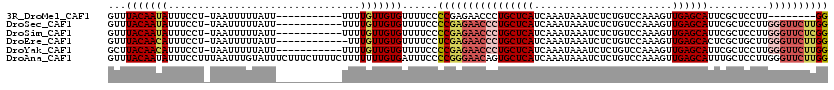
| Location | 2,669,434 – 2,669,534 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.76 |
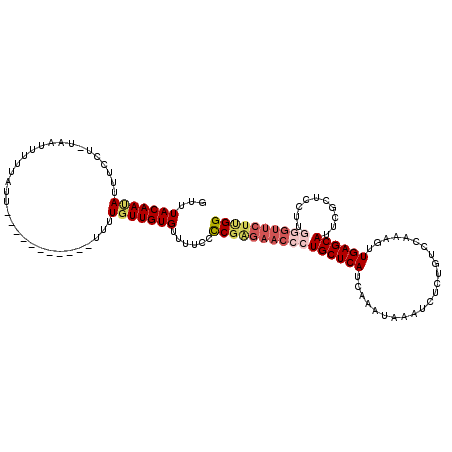
| Mean single sequence MFE | -22.02 |
| Consensus MFE | -17.83 |
| Energy contribution | -19.13 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.588592 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2669434 100 - 27905053 GUUUACAAUAUUUCCU-UAAUUUUUAUU-----------UUUUGUUGUGUUUUCCCCGAGAACCCUGCUCAUCAAAUAAAUCUCUGUCCAAAGUUGAGCAUUCGCUCCUU--------GG ...(((((((......-...........-----------...)))))))......(((((.....((((((.......................)))))).......)))--------)) ( -12.55) >DroSec_CAF1 28918 108 - 1 GUUUACAAUAUUUCCU-UAAUUUUUAUU-----------UUUUGUUGUGUUUUCCCCGAGAACCCUGCUCAUCAAAUAAAUCUCUGUCCAAAGUUGAGCAUUCGCUCCUUGGGUUCUUGG ...(((((((......-...........-----------...)))))))......((((((((((((((((.......................))))))..........)))))))))) ( -23.65) >DroSim_CAF1 21991 108 - 1 GUUUACAAUAUUUCCU-UAAUUUUUAUU-----------UUUUGUUGUGUUUUCCCCGAGAACCCUGCUCAUCAAAUAAAUCUCUGUCCAAAGUUGAGCAUUCGCUCCUUGGGUUCUCGG ...(((((((......-...........-----------...)))))))......((((((((((((((((.......................))))))..........)))))))))) ( -25.75) >DroEre_CAF1 21966 107 - 1 GUUUACAACAUUUCCU-UAAUUUUUAUU------------UUUGUUGUGUUUUCCUCGAGAACCCUGCUCAUCAAAUAAAUCUCUGUCCAAAGUUGAGCACUCGCUGCUUGGGUUCUUGG ...(((((((......-...........------------..))))))).......(((((((((((((((.......................))))))..........))))))))). ( -23.41) >DroYak_CAF1 29140 108 - 1 GCUUACAACAUUUCCU-UAAUUUUUAUU-----------UUUUGUUGUGUUUUCCCCGAGAACCCUGCUCAUCAAAUAAAUCUCUGUCCAAAGUUGAGCAUUCGCUCCUUGGGUUCUUGG ...(((((((......-...........-----------...)))))))......((((((((((((((((.......................))))))..........)))))))))) ( -25.55) >DroAna_CAF1 16954 120 - 1 GUUUACAAUAUUUCCUUUAAUUUGUAUUUCUUUCUUUUCUUUUUUUGUGAUUUCCCCGGGAACAGUGCUCAUCAAAUAAAUCUCUGUCCAAAGUUGAGCAUUUGCUCCUUGGGUUCUUGG ...(((((.(((......))))))))......................((...(((.((((.(((((((((.......................)))))).))).)))).))).)).... ( -21.20) >consensus GUUUACAAUAUUUCCU_UAAUUUUUAUU___________UUUUGUUGUGUUUUCCCCGAGAACCCUGCUCAUCAAAUAAAUCUCUGUCCAAAGUUGAGCAUUCGCUCCUUGGGUUCUUGG ...(((((((................................)))))))......((((((((((((((((.......................))))))..........)))))))))) (-17.83 = -19.13 + 1.31)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:51:30 2006