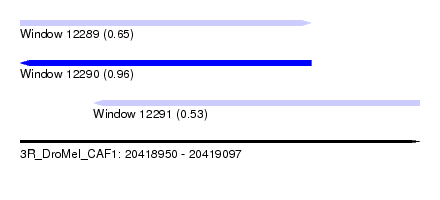
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 20,418,950 – 20,419,097 |
| Length | 147 |
| Max. P | 0.964577 |

| Location | 20,418,950 – 20,419,057 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 80.41 |
| Mean single sequence MFE | -17.07 |
| Consensus MFE | -14.97 |
| Energy contribution | -14.46 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.650245 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20418950 107 + 27905053 AAAUCAGAUUAUUUUCCCCCCCCUGGCUUUUAUCAGGGUUAAAUCAGCGUAAUUGUUUGGCCAUCUCCUUAUCAUUAUCAGACUGAAUCUAAUCAAAAGCCAAACAA ....................((((((......))))))..............(((((((((..(((...((....))..))).(((......)))...))))))))) ( -18.30) >DroSec_CAF1 2378 104 + 1 AAAUCAGAUUACUU---CCCCCCUGACUUUUAUCAGGGUUAAAUCAGCGUAAUUGUUUGGCCAUCUUCUUAUCGUUAUCAGCCUAGAUCUAAUCAAAAGCCAAACAA ......((((....---...((((((......))))))...)))).......(((((((((.........(((............)))..........))))))))) ( -19.21) >DroSim_CAF1 1947 104 + 1 AAAUCAGAUUAUUU---CCCCCCUGGCUUUUAUCAGGGUUAAAUCAGCGUAAUUGUUUGGCCAUCUUCUUAUCAUUAUCAGACUAGAUCUAAUCAAAAGCCAAACAA ......((((....---...((((((......))))))...)))).......(((((((((.....(((..((.......))..)))...........))))))))) ( -18.49) >DroYak_CAF1 1962 82 + 1 -----------GUU-----CACUUGGCUUUUAUCAGGGAUUAAUCAGCGUAAUUGUUUGGUCUUCU---------UAUCAGACUUUACCUAAUCAAAAACCAAACAA -----------...-----...((((.((((((.((((((((.......)))))(((((((.....---------.)))))))....))).)).)))).)))).... ( -12.30) >consensus AAAUCAGAUUAUUU___CCCCCCUGGCUUUUAUCAGGGUUAAAUCAGCGUAAUUGUUUGGCCAUCUUCUUAUCAUUAUCAGACUAGAUCUAAUCAAAAGCCAAACAA ....................((((((......))))))..............(((((((((.....................................))))))))) (-14.97 = -14.46 + -0.50)



| Location | 20,418,950 – 20,419,057 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 80.41 |
| Mean single sequence MFE | -25.23 |
| Consensus MFE | -19.73 |
| Energy contribution | -19.98 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.964577 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20418950 107 - 27905053 UUGUUUGGCUUUUGAUUAGAUUCAGUCUGAUAAUGAUAAGGAGAUGGCCAAACAAUUACGCUGAUUUAACCCUGAUAAAAGCCAGGGGGGGGAAAAUAAUCUGAUUU ((((((((((.((.(((((((...)))))))...........)).)))))))))).........(((..(((((........)))))..)))............... ( -25.00) >DroSec_CAF1 2378 104 - 1 UUGUUUGGCUUUUGAUUAGAUCUAGGCUGAUAACGAUAAGAAGAUGGCCAAACAAUUACGCUGAUUUAACCCUGAUAAAAGUCAGGGGGG---AAGUAAUCUGAUUU (((((((((((((..(((.(((......))).....)))..))).)))))))))).........(((..(((((((....)))))))..)---))............ ( -28.50) >DroSim_CAF1 1947 104 - 1 UUGUUUGGCUUUUGAUUAGAUCUAGUCUGAUAAUGAUAAGAAGAUGGCCAAACAAUUACGCUGAUUUAACCCUGAUAAAAGCCAGGGGGG---AAAUAAUCUGAUUU ((((((((((.((.(((((((...)))))))...........)).)))))))))).........(((..(((((........)))))..)---))............ ( -26.20) >DroYak_CAF1 1962 82 - 1 UUGUUUGGUUUUUGAUUAGGUAAAGUCUGAUA---------AGAAGACCAAACAAUUACGCUGAUUAAUCCCUGAUAAAAGCCAAGUG-----AAC----------- (((((((((((((.((((((.....)))))).---------.)))))))))))))(((((((.((((.....))))...)))...)))-----)..----------- ( -21.20) >consensus UUGUUUGGCUUUUGAUUAGAUCUAGUCUGAUAAUGAUAAGAAGAUGGCCAAACAAUUACGCUGAUUUAACCCUGAUAAAAGCCAGGGGGG___AAAUAAUCUGAUUU ((((((((((.((.((((((.....))))))...........)).))))))))))..............(((((........))))).................... (-19.73 = -19.98 + 0.25)


| Location | 20,418,977 – 20,419,097 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.50 |
| Mean single sequence MFE | -23.60 |
| Consensus MFE | -18.40 |
| Energy contribution | -18.20 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.78 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.532003 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20418977 120 - 27905053 CUAGCAAAGUUCUCAUUGCCUUCUCAACAAAUAUAGCAAAUUGUUUGGCUUUUGAUUAGAUUCAGUCUGAUAAUGAUAAGGAGAUGGCCAAACAAUUACGCUGAUUUAACCCUGAUAAAA ...((((.(....).)))).........((((.((((.((((((((((((.((.(((((((...)))))))...........)).))))))))))))..))))))))............. ( -24.30) >DroSec_CAF1 2402 120 - 1 CCAGCAAAGUUCCCAUUGCCUUCUCAACAAAUAUAGCAAAUUGUUUGGCUUUUGAUUAGAUCUAGGCUGAUAACGAUAAGAAGAUGGCCAAACAAUUACGCUGAUUUAACCCUGAUAAAA .((((...........(((................)))(((((((((((((((..(((.(((......))).....)))..))).))))))))))))..))))................. ( -24.09) >DroSim_CAF1 1971 120 - 1 CCAGCAAAGUUCUCAUUGCCUUCUCAACAAAUAUAGGAAAUUGUUUGGCUUUUGAUUAGAUCUAGUCUGAUAAUGAUAAGAAGAUGGCCAAACAAUUACGCUGAUUUAACCCUGAUAAAA .((((.............(((.............))).((((((((((((.((.(((((((...)))))))...........)).))))))))))))..))))................. ( -25.32) >DroEre_CAF1 1919 109 - 1 CGAGCA-AGUUCUAUAUGCAGUCUCAGCAAAUGUUGGAAAUUGUUUGACUUUUGAUUAGGUCUAGUCUGAUA---------AGAAGGCCAAACAAUUACGCUGAUUUAUCCAUGA-AAAA ..(((.-..............((.((((....))))))(((((((((.(((((.((((((.....)))))).---------)))))..)))))))))..))).............-.... ( -23.30) >DroYak_CAF1 1973 98 - 1 CUAGUA-AGUUCUAUAUGCAAUCUCA------------AAUUGUUUGGUUUUUGAUUAGGUAAAGUCUGAUA---------AGAAGACCAAACAAUUACGCUGAUUAAUCCCUGAUAAAA .((((.-.((.......)).......------------(((((((((((((((.((((((.....)))))).---------.)))))))))))))))..))))................. ( -21.00) >consensus CCAGCAAAGUUCUCAUUGCCUUCUCAACAAAUAUAGCAAAUUGUUUGGCUUUUGAUUAGAUCUAGUCUGAUAA_GAUAAG_AGAUGGCCAAACAAUUACGCUGAUUUAACCCUGAUAAAA .((((...((.......))...................((((((((((((.((.((((((.....))))))...........)).))))))))))))..))))................. (-18.40 = -18.20 + -0.20)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:50:17 2006