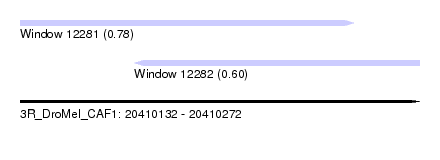
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 20,410,132 – 20,410,272 |
| Length | 140 |
| Max. P | 0.783799 |

| Location | 20,410,132 – 20,410,249 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 76.41 |
| Mean single sequence MFE | -45.20 |
| Consensus MFE | -23.68 |
| Energy contribution | -24.93 |
| Covariance contribution | 1.26 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.783799 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20410132 117 + 27905053 UUCUGACUGGCGAUUGAUUGCCAUUCCUGGCCUUUCGGGGUGGACUCUGAUCGGAAUCCCUGUGCUUUUGGCGUGGCGGAGACUGAUCGCCAUCUCGCCCUCGUCUGGUGGGACUCU ....((.((((((....)))))).))((.(((...(((((((((...(((((((..((((..(((.....)))..).)))..)))))))...)).))))).))...))).))..... ( -46.90) >DroVir_CAF1 41058 114 + 1 UCCUGGGCGGCGACUGAUCCGCAUCUCUUCUUCUCCUGGGCGGCGACUGAUCCACAUCACGACC---CCGCCUGGGCGGGCUUUGAUCUGCCCCACGCCGUCGUCUGACUGGACUAU .....(((((((......................((..(((((.(.(((((....)))).).).---)))))..)).((((........))))..)))))))((((....))))... ( -42.50) >DroSec_CAF1 32517 117 + 1 UUCCGGCUGGCGAUGGUUUGUCAUUCCUGGCCUUUCUGGGCGGACUCUGAUCGGAAUCUCUCCGCUUUCUGCGAGACGGAGACUGAUCGCCAUCUCGCCCUCGUCUAGGAGGACUCU (((((((..(.(((((....))))).)..))).....(((((((...(((((((..(((.(((((.....))).)).)))..)))))))...)).))))).......))))...... ( -44.90) >DroSim_CAF1 35822 117 + 1 UCCUUGCUGGCGAAUGUUUGUCAUUCCUGGCCUUUCUGGGCGGACUCUGAUCGGAAUCUCUCCGCUUUCUGCGAGGCGGAGACUGAUCGCCAUCUCGCCCUCGUCUAGGAGGACUCU (((((((..(.(((((.....))))))..))......(((((((...(((((((....((((((((((....)))))))))))))))))...)).)))))........))))).... ( -48.70) >DroEre_CAF1 32124 117 + 1 UCCUGGCUGGCGACUGAUUGCCAUUCCUGGCUUUUCUGGGUGGACUCUGAUCGGAAUCUCUGCGCUUUCGGCGAGGCGGCGAUUGAUCGCCAACUCUUUCUCGCCUUGGAGGACUCU (((.(((((((((..(((((((...(((.(((.....(.(..((.(((....)))...))..).)....))).))).)))))))..))))))..........)))..)))....... ( -41.30) >DroYak_CAF1 36449 117 + 1 UCCUAGCUGGCGAUUGAUCGCCAUUUUUGGCUUUUCUGGGUGGACUCUGAUCGGAAUCUCUGCGCUUUCGGCCAGGCGGCGAUUGAUCGCCAUCUCUUCCCCGUCUGGGCGGACUCU ....((.(((((((..((((((...(((((((.....(.(..((.(((....)))...))..).)....))))))).))))))..))))))).)).....((((....))))..... ( -46.90) >consensus UCCUGGCUGGCGAUUGAUUGCCAUUCCUGGCCUUUCUGGGCGGACUCUGAUCGGAAUCUCUGCGCUUUCGGCGAGGCGGAGACUGAUCGCCAUCUCGCCCUCGUCUGGGAGGACUCU (((.....(((((......((((....))))......(((((((...(((((((..(((((.(((.....))).)).)))..)))))))...)).)))))))))).....))).... (-23.68 = -24.93 + 1.26)


| Location | 20,410,172 – 20,410,272 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 73.88 |
| Mean single sequence MFE | -37.81 |
| Consensus MFE | -15.58 |
| Energy contribution | -17.13 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.41 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.595492 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20410172 100 - 27905053 -----------------AAGAAGGGCGGGAAGCUCAGAUCAGAGUCCCACCAGACGAGGGCGAGAUGGCGAUCAGUCUCCGCCACGCCAAAAGCACAGGGAUUCCGAUCAGAGUCCA -----------------.....(((((((...))).((((.(((((((..........((((.((.(((.....)))))))))..((.....))...))))))).))))...)))). ( -36.90) >DroVir_CAF1 41098 112 - 1 CAAUCAAAGCCCUCACAGACAACGCCGCG--ACUCAGAUCAUAGUCCAGUCAGACGACGGCGUGGGGCAGAUCAAAGCCCGCCCAGGCGG---GGUCGUGAUGUGGAUCAGUCGCCG ........(((((........((((((.(--(((........))))).(((....))))))))))))).........(((((....))))---)...(((((........))))).. ( -37.20) >DroSec_CAF1 32557 100 - 1 -----------------AAGAAGGGCGGCAAGCUCAGAUCAGAGUCCUCCUAGACGAGGGCGAGAUGGCGAUCAGUCUCCGUCUCGCAGAAAGCGGAGAGAUUCCGAUCAGAGUCCG -----------------........(((...((((.((((.(((((((((.........(((((((((.((....)).))))))))).......)))).))))).)))).))))))) ( -40.99) >DroSim_CAF1 35862 100 - 1 -----------------AAGAAGGGCGGCAAGCUCAGAUCAGAGUCCUCCUAGACGAGGGCGAGAUGGCGAUCAGUCUCCGCCUCGCAGAAAGCGGAGAGAUUCCGAUCAGAGUCCG -----------------........(((...((((.((((.(((((((((.....((((.((.((.(((.....)))))))))))((.....)))))).))))).)))).))))))) ( -38.50) >DroEre_CAF1 32164 100 - 1 -----------------AAGAAGGGCGGAAAGCUCAGAUCAGAGUCCUCCAAGGCGAGAAAGAGUUGGCGAUCAAUCGCCGCCUCGCCGAAAGCGCAGAGAUUCCGAUCAGAGUCCA -----------------.....(((((((..((((......))))..)))..((((.(((.(((.((((((....)))))).)))(((....).)).....))))).))...)))). ( -35.80) >DroYak_CAF1 36489 100 - 1 -----------------AAGAAGGGCGGAAAGCUCGGAUCAGAGUCCGCCCAGACGGGGAAGAGAUGGCGAUCAAUCGCCGCCUGGCCGAAAGCGCAGAGAUUCCGAUCAGAGUCCA -----------------.....((((((...((((......)))))))))).(((((((...((.((((((....)))))).)).(((....).)).....))))).))........ ( -37.50) >consensus _________________AAGAAGGGCGGAAAGCUCAGAUCAGAGUCCUCCCAGACGAGGGCGAGAUGGCGAUCAAUCUCCGCCUCGCCGAAAGCGCAGAGAUUCCGAUCAGAGUCCA ..........................((...((((.((((.((((((...................((((.........))))..((.....))...).))))).)))).)))))). (-15.58 = -17.13 + 1.56)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:50:09 2006