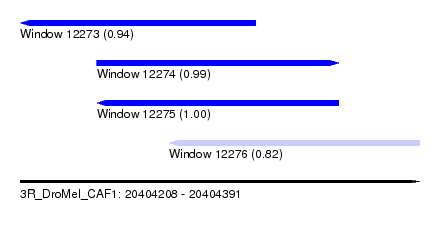
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 20,404,208 – 20,404,391 |
| Length | 183 |
| Max. P | 0.999889 |

| Location | 20,404,208 – 20,404,316 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 95.02 |
| Mean single sequence MFE | -31.33 |
| Consensus MFE | -25.98 |
| Energy contribution | -26.38 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.942675 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20404208 108 - 27905053 ACGGAACCGAAUGGAUCGGAUCUUCGAAGAGGGUUGCUGCACCCCAUUAAAAGAAACUGCAUGCGGCAACCCACAGUUAAGUAUGUAAUUUUCUGCCAUC-CAUAUGCA .((....)).((((((.((......((((((((((((((((....................)))))))))))(((........)))..)))))..)))))-)))..... ( -30.65) >DroSec_CAF1 26783 108 - 1 ACGGAACCGAAUGGAUCGGAUCUUCGAAGAGGGUUGCUGCACCCCAUUAUAAGAAACUGCAUGCGGCAACCCACAGUUAAGUAUGUAAUUUUCUGCCAUC-CAUAUGCA .((....)).((((((.((......((((((((((((((((....................)))))))))))(((........)))..)))))..)))))-)))..... ( -30.65) >DroSim_CAF1 29784 108 - 1 ACGGAACCGAAUGGAUCGGAUCUUCGAAGAGGGUUGCUGCACCCCAUUAAAAGAAACUGCAUGCGGCAACCCACAGUUAAGUAUGCAAUUUUCUGCCAUC-CAUAUGCA .((....)).((((((.((...........(((((((((((....................))))))))))).(((..((........))..))))))))-)))..... ( -30.25) >DroEre_CAF1 26388 109 - 1 ACGGAACCGAAUGGAUCGGAUCUUCCGGGAGGGUUGCUGCAACCCAUUAUAAGAAACUGCAUGCGGCAACCCACAGUUAAGUAUGUAAUUUUCUGCCAUCCCAUAUGCA ..((((((((.....))))...))))(((((((((((((((....................))))))))))).(((..((........))..)))...))))....... ( -35.05) >DroYak_CAF1 30578 108 - 1 ACGGAACCGAAUGAAUCGGAUCUUCCGGCAGGGUAGCUGCAAUCCAUUAUAAGAAACUGCAUGCGGCAACCCACAGUUAAGUAUGUAAUUUUCUGCCAUC-CAUAUGCA ..(((.((((.....)))).......((((((((.((((((....................)))))).))))(((........))).......)))).))-)....... ( -30.05) >consensus ACGGAACCGAAUGGAUCGGAUCUUCGAAGAGGGUUGCUGCACCCCAUUAUAAGAAACUGCAUGCGGCAACCCACAGUUAAGUAUGUAAUUUUCUGCCAUC_CAUAUGCA ..(((.((((.....)))).))).......(((((((((((....................)))))))))))........((((((................)))))). (-25.98 = -26.38 + 0.40)

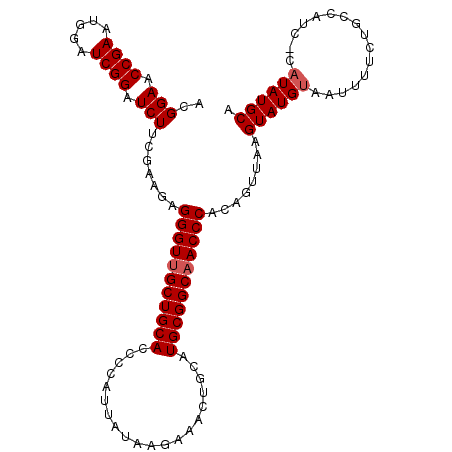
| Location | 20,404,243 – 20,404,354 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 94.23 |
| Mean single sequence MFE | -33.18 |
| Consensus MFE | -28.92 |
| Energy contribution | -29.72 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.87 |
| SVM decision value | 2.33 |
| SVM RNA-class probability | 0.992496 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20404243 111 + 27905053 UGGGUUGCCGCAUGCAGUUUCUUUUAAUGGGGUGCAGCAACCCUCUUCGAAGAUCCGAUCCAUUCGGUUCCGUUUCGAUACGAUUGCUUUUUCCACGCGAAAAACUCAUUU .(((((((.((((.(.(((......))).).)))).)))))))...((((((..((((.....)))).....)))))).....((((.........))))........... ( -32.20) >DroSec_CAF1 26818 111 + 1 UGGGUUGCCGCAUGCAGUUUCUUAUAAUGGGGUGCAGCAACCCUCUUCGAAGAUCCGAUCCAUUCGGUUCCGUUUCGAUACGAUUGCUUUUUCCACGCGGAAAACUCAUUU .(((((((.((((.(.(((......))).).)))).)))))))...((((((..((((.....)))).....))))))......((..((((((....))))))..))... ( -35.60) >DroSim_CAF1 29819 111 + 1 UGGGUUGCCGCAUGCAGUUUCUUUUAAUGGGGUGCAGCAACCCUCUUCGAAGAUCCGAUCCAUUCGGUUCCGUUUCGAUACGAUUGCUUUUUCCACGCGGAAAACUCAUUU .(((((((.((((.(.(((......))).).)))).)))))))...((((((..((((.....)))).....))))))......((..((((((....))))))..))... ( -35.60) >DroEre_CAF1 26424 111 + 1 UGGGUUGCCGCAUGCAGUUUCUUAUAAUGGGUUGCAGCAACCCUCCCGGAAGAUCCGAUCCAUUCGGUUCCGUUUCGAUACGAUUGCUUUUUCCACACGGAAAACUCAUUU .(((((((.(((.((.(((......)))..))))).)))))))(((.((((((..(((((...((((.......))))...)))))..))))))....))).......... ( -33.50) >DroYak_CAF1 30613 111 + 1 UGGGUUGCCGCAUGCAGUUUCUUAUAAUGGAUUGCAGCUACCCUGCCGGAAGAUCCGAUUCAUUCGGUUCCGUUUCGAUACGACUGCUUUUUCCCCACGGAAAACUCAUUU ((((((.......((((((......((((((..((((.....))))(....)..((((.....)))).)))))).......))))))...((((....))))))))))... ( -29.02) >consensus UGGGUUGCCGCAUGCAGUUUCUUAUAAUGGGGUGCAGCAACCCUCUUCGAAGAUCCGAUCCAUUCGGUUCCGUUUCGAUACGAUUGCUUUUUCCACGCGGAAAACUCAUUU .(((((((.((((.(.(((......))).).)))).)))))))........((.((((.....)))).))(((......)))..((..((((((....))))))..))... (-28.92 = -29.72 + 0.80)

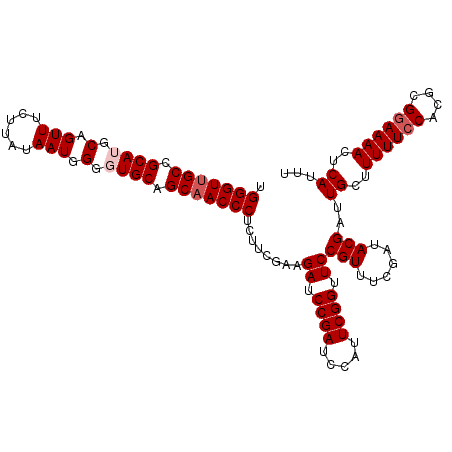

| Location | 20,404,243 – 20,404,354 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 94.23 |
| Mean single sequence MFE | -34.81 |
| Consensus MFE | -32.73 |
| Energy contribution | -32.93 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.94 |
| SVM decision value | 4.40 |
| SVM RNA-class probability | 0.999889 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20404243 111 - 27905053 AAAUGAGUUUUUCGCGUGGAAAAAGCAAUCGUAUCGAAACGGAACCGAAUGGAUCGGAUCUUCGAAGAGGGUUGCUGCACCCCAUUAAAAGAAACUGCAUGCGGCAACCCA ...((..((((((....))))))..)).((...((((...(((.((((.....)))).))))))).))(((((((((((....................))))))))))). ( -33.05) >DroSec_CAF1 26818 111 - 1 AAAUGAGUUUUCCGCGUGGAAAAAGCAAUCGUAUCGAAACGGAACCGAAUGGAUCGGAUCUUCGAAGAGGGUUGCUGCACCCCAUUAUAAGAAACUGCAUGCGGCAACCCA ...((..((((((....))))))..)).((...((((...(((.((((.....)))).))))))).))(((((((((((....................))))))))))). ( -35.95) >DroSim_CAF1 29819 111 - 1 AAAUGAGUUUUCCGCGUGGAAAAAGCAAUCGUAUCGAAACGGAACCGAAUGGAUCGGAUCUUCGAAGAGGGUUGCUGCACCCCAUUAAAAGAAACUGCAUGCGGCAACCCA ...((..((((((....))))))..)).((...((((...(((.((((.....)))).))))))).))(((((((((((....................))))))))))). ( -35.95) >DroEre_CAF1 26424 111 - 1 AAAUGAGUUUUCCGUGUGGAAAAAGCAAUCGUAUCGAAACGGAACCGAAUGGAUCGGAUCUUCCGGGAGGGUUGCUGCAACCCAUUAUAAGAAACUGCAUGCGGCAACCCA ...((..((((((....))))))..))............(((((((((.....))))...)))))...(((((((((((....................))))))))))). ( -36.15) >DroYak_CAF1 30613 111 - 1 AAAUGAGUUUUCCGUGGGGAAAAAGCAGUCGUAUCGAAACGGAACCGAAUGAAUCGGAUCUUCCGGCAGGGUAGCUGCAAUCCAUUAUAAGAAACUGCAUGCGGCAACCCA ...((..((((((....))))))..))(((((......))((((((((.....))))...))))))).((((.((((((....................)))))).)))). ( -32.95) >consensus AAAUGAGUUUUCCGCGUGGAAAAAGCAAUCGUAUCGAAACGGAACCGAAUGGAUCGGAUCUUCGAAGAGGGUUGCUGCACCCCAUUAUAAGAAACUGCAUGCGGCAACCCA ...((..((((((....))))))..))......((((...(((.((((.....)))).)))))))...(((((((((((....................))))))))))). (-32.73 = -32.93 + 0.20)

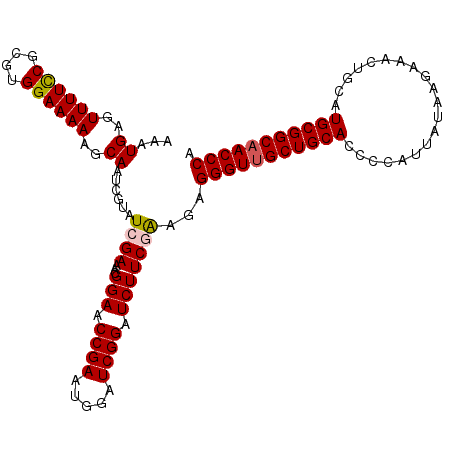
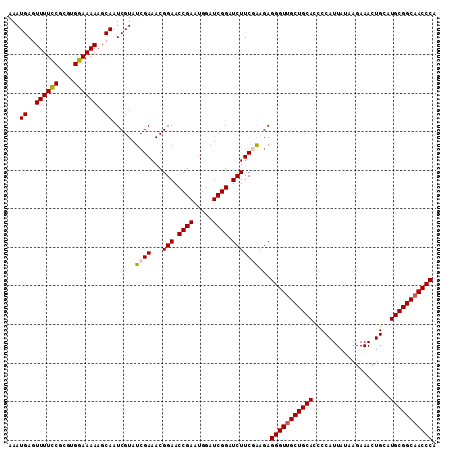
| Location | 20,404,276 – 20,404,391 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 95.15 |
| Mean single sequence MFE | -25.84 |
| Consensus MFE | -23.36 |
| Energy contribution | -23.40 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.818020 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20404276 115 - 27905053 CAAUAGAAAUGUAAAUAGGA-AAUUUUAAUAAUGAUUGAAAUGAGUUUUUCGCGUGGAAAAAGCAAUCGUAUCGAAACGGAACCGAAUGGAUCGGAUCUUCGAAGAGGGUUGCUGC ........(((..((.....-.((((((((....))))))))......))..)))......(((((((.(.((((...(((.((((.....)))).)))))))..).))))))).. ( -24.60) >DroSec_CAF1 26851 115 - 1 CAAUAGAAAUGUAAAUAGGA-AAUUUUAAUAAUGAUUGAAAUGAGUUUUCCGCGUGGAAAAAGCAAUCGUAUCGAAACGGAACCGAAUGGAUCGGAUCUUCGAAGAGGGUUGCUGC ..(((....)))........-.((((((((....))))))))...((((((....))))))(((((((.(.((((...(((.((((.....)))).)))))))..).))))))).. ( -26.50) >DroSim_CAF1 29852 115 - 1 CAAUAGAAAUGUAAAUAGGA-AAUUUUAAUAAUGAUUGAAAUGAGUUUUCCGCGUGGAAAAAGCAAUCGUAUCGAAACGGAACCGAAUGGAUCGGAUCUUCGAAGAGGGUUGCUGC ..(((....)))........-.((((((((....))))))))...((((((....))))))(((((((.(.((((...(((.((((.....)))).)))))))..).))))))).. ( -26.50) >DroEre_CAF1 26457 116 - 1 CAAUAGAAAUGUAAAUAGGAAAAUUUUAAUAAUGAUUGAAAUGAGUUUUCCGUGUGGAAAAAGCAAUCGUAUCGAAACGGAACCGAAUGGAUCGGAUCUUCCGGGAGGGUUGCUGC ..(((....))).....(((((((((................)))))))))..........(((((((...((....(((((((((.....))))...))))).)).))))))).. ( -25.69) >DroYak_CAF1 30646 115 - 1 CAAUAGAAAUGUAAAUAGCA-AAUUUUAAUAAUGAUUGAAAUGAGUUUUCCGUGGGGAAAAAGCAGUCGUAUCGAAACGGAACCGAAUGAAUCGGAUCUUCCGGCAGGGUAGCUGC ..(((....)))...((((.-..(((..(((.((((((.......((((((....))))))..)))))))))..)))(((((((((.....))))...)))))........)))). ( -25.90) >consensus CAAUAGAAAUGUAAAUAGGA_AAUUUUAAUAAUGAUUGAAAUGAGUUUUCCGCGUGGAAAAAGCAAUCGUAUCGAAACGGAACCGAAUGGAUCGGAUCUUCGAAGAGGGUUGCUGC ..(((....)))..........((((((((....))))))))...((((((....))))))(((((((.(.((((...(((.((((.....)))).)))))))..).))))))).. (-23.36 = -23.40 + 0.04)

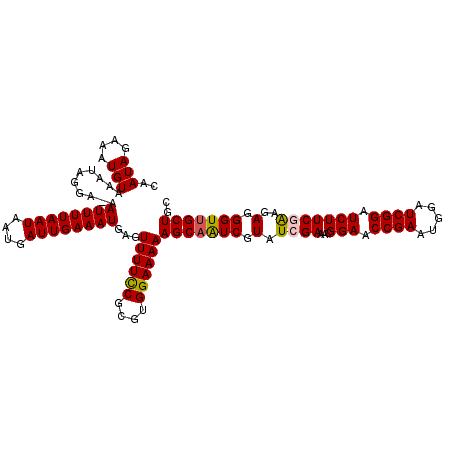
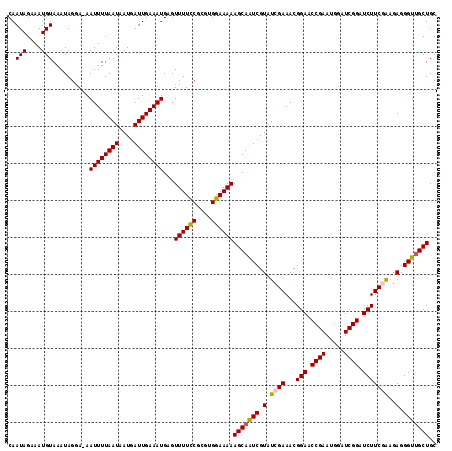
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:50:03 2006