
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 20,392,704 – 20,392,859 |
| Length | 155 |
| Max. P | 0.868969 |

| Location | 20,392,704 – 20,392,819 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 79.90 |
| Mean single sequence MFE | -25.35 |
| Consensus MFE | -16.57 |
| Energy contribution | -17.58 |
| Covariance contribution | 1.01 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.561613 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
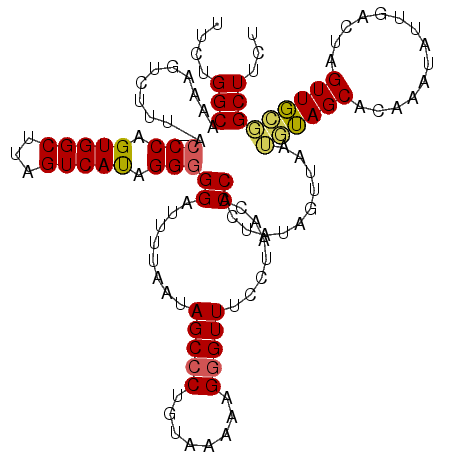
>3R_DroMel_CAF1 20392704 115 + 27905053 CACCGUCUGUCU--CUUUUGCUUUGAUAUUCCCCUUCUUUCUGGCAAAAGUCUUU-ACCCAGUGGCUUAGUCAUGGGGGGUUUUUAAUAGCCCUGUAAAAUGGGUUUCUUUACCCACU ............--((((((((..((.............)).)))))))).....-.((((.((((...))))))))((((.......(((((........))))).....))))... ( -28.92) >DroSec_CAF1 15333 115 + 1 CGUCUUCUGUCU--CUCUUGCUUUGAUAUUCCCUUUCUUUCUGGCAAAAGUCUUU-ACCCAGUGGCUUAGUCACAGGUGGAUUUUAAUAGCCCUGUAAUAAGGGUUUCCUAACCCACU ......((((..--((.(((((..((.............)).)))))(((((...-.......)))))))..))))((((.((.....((((((......))))))....)).)))). ( -24.12) >DroSim_CAF1 18339 115 + 1 CGUCUUCUGUCU--CUUUUGCUUUGAUAUUCCCUUUCUUUCUGGCAAAAGUCUUU-UCCCAGUGGCUUAGUCACAGGGGGAUUUUAAUAGCCCUGUAAAGAGAGUUUCCUAACCCCCU .........(((--((((.(((((((.((((((((......(((.(((.....))-).)))(((((...))))))))))))).)))).))).....)))))))............... ( -25.80) >DroEre_CAF1 14806 94 + 1 CACUCUGUCUCU--CUUUUGCUGUGAUAUUCCCUUUCUUUCUGGCAAAAGACUUUUGCCCAGUGGCUUAGUCAUAGGGGGAUUU----------------------UAAUAACCCACU ......((((((--(((((((((.((.............))))))))))((((...(((....)))..))))...)))))))..----------------------............ ( -21.92) >DroYak_CAF1 18791 117 + 1 CACUCUGUCUCUCUCUCUCGCUUUGAUAUUCCAUUUCUUUCUGGCAAAAGUCUUUUGCCCAGUGGCUUAGUCAUAGGGGGACUU-AAUAGCCCUGUAAAAUGGGUUUCCUAACCCACC ......((((((((............................((((((.....))))))..(((((...)))))))))))))..-...............((((((....)))))).. ( -26.00) >consensus CACCCUCUGUCU__CUUUUGCUUUGAUAUUCCCUUUCUUUCUGGCAAAAGUCUUU_ACCCAGUGGCUUAGUCAUAGGGGGAUUUUAAUAGCCCUGUAAAAAGGGUUUCCUAACCCACU ..............((((((((..((.............)).)))))))).......(((.(((((...))))).)))((........(((((........))))).......))... (-16.57 = -17.58 + 1.01)



| Location | 20,392,740 – 20,392,859 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.22 |
| Mean single sequence MFE | -29.68 |
| Consensus MFE | -14.13 |
| Energy contribution | -14.42 |
| Covariance contribution | 0.29 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.868969 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20392740 119 + 27905053 UUCUGGCAAAAGUCUUU-ACCCAGUGGCUUAGUCAUGGGGGGUUUUUAAUAGCCCUGUAAAAUGGGUUUCUUUACCCACUAUAGUUAAUAUAGCACAAAUAUUGACUAGUUGUGGCUUCU ........((((.....-((((((((((...)))))(.((((((......)))))).)....)))))..))))..((((..((((((((((.......))))))))))...))))..... ( -32.60) >DroSec_CAF1 15369 119 + 1 UUCUGGCAAAAGUCUUU-ACCCAGUGGCUUAGUCACAGGUGGAUUUUAAUAGCCCUGUAAUAAGGGUUUCCUAACCCACUAUAGUUAAUGUAGCACAAAUAUUGACUAGUUGCGGCUUUU ....(((.(((((((..-(((..(((((...))))).))))))))))....)))(((((((..(((((....)))))....((((((((((.......)))))))))))))))))..... ( -34.20) >DroSim_CAF1 18375 119 + 1 UUCUGGCAAAAGUCUUU-UCCCAGUGGCUUAGUCACAGGGGGAUUUUAAUAGCCCUGUAAAGAGAGUUUCCUAACCCCCUAUAGUUAAUGUAGCACAAAUAUUGACUAGUUGCGGCUUUU .......(((((((...-.....(((((...)))))((((((.((.....(((.((......)).)))....)))))))).((((((((((.......)))))))))).....))))))) ( -29.50) >DroEre_CAF1 14842 97 + 1 UUCUGGCAAAAGACUUUUGCCCAGUGGCUUAGUCAUAGGGGGAUUU----------------------UAAUAACCCACUAUUAUUAAAGGAGCACA-AUACUGACAAGUUUCUGCUUCU ..((((((((.....)))).))))....(((((.(((((((.....----------------------......))).)))).)))))(((((((..-..(((....)))...))))))) ( -24.70) >DroYak_CAF1 18829 95 + 1 UUCUGGCAAAAGUCUUUUGCCCAGUGGCUUAGUCAUAGGGGGACUU-AAUAGCCCUGUAAAAUGGGUUUCCUAACCCACCCAUGUUCAAG------------------------GCUUCU ....((((((.....)))))).((.(((((.(.(((.(((((.((.-...))))).......((((((....)))))))).)))..).))------------------------))).)) ( -27.40) >consensus UUCUGGCAAAAGUCUUU_ACCCAGUGGCUUAGUCAUAGGGGGAUUUUAAUAGCCCUGUAAAAAGGGUUUCCUAACCCACUAUAGUUAAUGUAGCACAAAUAUUGACUAGUUGCGGCUUCU ....(((............(((.(((((...))))).)))((........(((((........))))).......))...........((((((..............)))))))))... (-14.13 = -14.42 + 0.29)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:49:56 2006