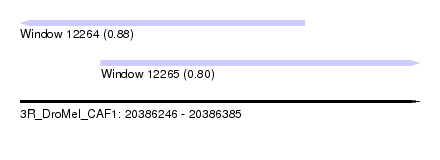
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 20,386,246 – 20,386,385 |
| Length | 139 |
| Max. P | 0.876596 |

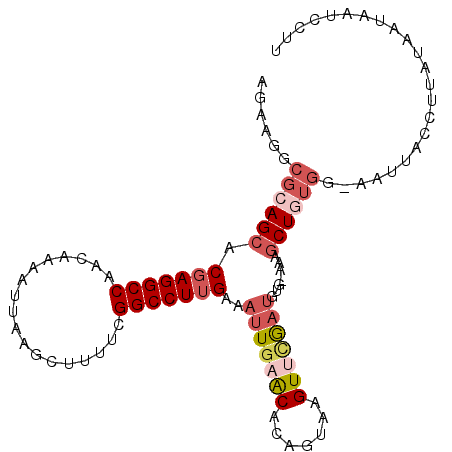
| Location | 20,386,246 – 20,386,345 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 81.43 |
| Mean single sequence MFE | -27.19 |
| Consensus MFE | -19.11 |
| Energy contribution | -20.47 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.876596 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20386246 99 - 27905053 AGAAGGCGCAGCACGAGGCCAACAAAAUUAAGUUGUUCGGCCUUGAAAUUGAACACAGUGAGUUCAAUGUG-AAAGCUGUGG-AAAUAUCUUAUAA---UCCUU ..((((((((((.((((((((((((.......))))).)))))))..(((((((.......)))))))...-...))))))(-(....))......---.)))) ( -31.60) >DroSec_CAF1 9137 102 - 1 AGAAGGCGCAGCACGAGGCCAACAAAAUUAAGCUUUUCGGCCUUGAAAUUGAACACAGUAAGUUCGAUGUG-AAUGCUUUGG-CAUUACCUUAUAAUAAUGCAU ..((((((..((.(((((((..................)))))))..(((((((.......))))))))).-..)))))).(-(((((........)))))).. ( -27.17) >DroSim_CAF1 12083 102 - 1 AGAAGGCGCAGCACGAGGCCAACAAAAUUAAGCUUUUCGGCCUUGAAAUUGAACACAGUAAGUUUGAUGUG-AAAGCUGUGG-CAUUACCUUAUAAUAAUGCAU ......((((((.(((((((..................)))))))..((..(((.......)))..))...-...))))))(-(((((........)))))).. ( -29.27) >DroEre_CAF1 8605 102 - 1 AGAAGGCGCAGCACGAGGCCAACAAAAUUAAGCUUUUCGGCCUUGAAAUUGAACACAGUAAGUGCGAUGCG-CAAGCUGUGG-AAUUACCUUAAAGUAAUCUUU .((((.((((((.(((((((..................)))))))................((((...)))-)..)))))).-.(((((......))))))))) ( -26.47) >DroYak_CAF1 12533 102 - 1 AGAAGGCGCAGCACGAGGCCAAUAAAAUUAAGCUAUUUGGCCUUGAAAUUGAACACAGUAAGUUGGAUGUG-CAAGCUGUGG-AAUUACCAUAUAAUAAUCCUU ..((((((((((.(((((((((..............)))))))))..(((.(((.......))).)))...-...)))))).-.((((.....))))...)))) ( -26.34) >DroMoj_CAF1 28188 100 - 1 AGAAGGCCCAGCACGAGGCCAACAAAAUCAAACUAUAUGGCCUGGAAAUUGAGCACAGUGAGUGCA---UGCAAUACUAUAUUGAUG-UAAUUGAACUAUAUCA ..........((((.((((((................)))))).).......((((.....)))).---)))..........(((((-((.......))))))) ( -22.29) >consensus AGAAGGCGCAGCACGAGGCCAACAAAAUUAAGCUUUUCGGCCUUGAAAUUGAACACAGUAAGUUCGAUGUG_AAAGCUGUGG_AAUUACCUUAUAAUAAUCCUU ......((((((.(((((((..................)))))))..(((((((.......))))))).......))))))....................... (-19.11 = -20.47 + 1.36)


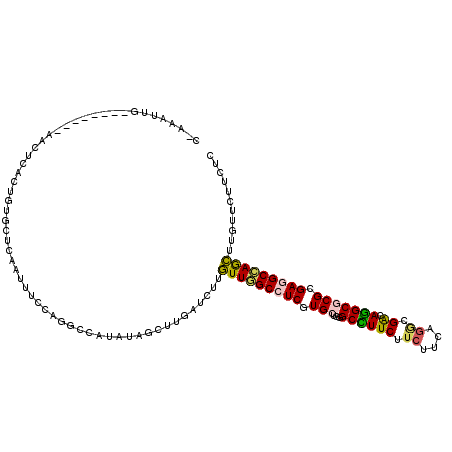
| Location | 20,386,274 – 20,386,385 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.70 |
| Mean single sequence MFE | -31.67 |
| Consensus MFE | -20.55 |
| Energy contribution | -20.22 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.802921 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20386274 111 + 27905053 C-ACAUUG--------AACUCACUGUGUUCAAUUUCAAGGCCGAACAACUUAAUUUUGUUGGCCUCGUGCUGCGCCUUCUUCUUCAGGCGACAGGCGCGUGAGGCCAGCUUGUUCUUCUC .-..((((--------(((.......))))))).........((((((.........((((((((((((((.(((((........)))))...)))))..)))))))))))))))..... ( -41.10) >DroVir_CAF1 10806 110 + 1 C-AAC-UC--------GACUCACUGUGCUCAAUUUCCAACCCAUAUAGUUUGAUCUUAUUAGCCUCAUGCUGGGCCUUCUUCUUCAGACGGCAGGCGCGAGAGGCUAAUUUGUUCUUCUC .-...-..--------.....((((((................))))))..((.(..(((((((((.((((..(((.(((.....))).))).))))...)))))))))..).))..... ( -26.49) >DroGri_CAF1 30407 111 + 1 C-AACUUC--------GACUCACUGUGCUCAAUUUCCAGGCCAUAGAGCUUGAUCUUGUUGGCCUCGUGCUGGGCCUUCUUCUUCAGGCGACAGGCGCGCGAGGCCAGCUUAUUCUUCUC .-....((--------(((((..((.(((.........)))))..))).))))....(((((((((((((...((((((.((....)).)).)))))))))))))))))........... ( -39.10) >DroWil_CAF1 12313 112 + 1 CUUUUUUG--------AACUUACUAUGUUCAAUUUCCAGUCCAUAUAGCUUUAUCUUGUUGGCCUCAUGCUGGGCUUUCUUCUUCAGACGACAAGCACGCGAUGCCAGCUUAUUCUUCUC .....(((--------(((.......)))))).........................((((((.((..((...((((((.((....)).)).))))..)))).))))))........... ( -22.00) >DroMoj_CAF1 28220 108 + 1 C-A---UG--------CACUCACUGUGCUCAAUUUCCAGGCCAUAUAGUUUGAUUUUGUUGGCCUCGUGCUGGGCCUUCUUCUUCAGCCGGCAGGCACGCGAUGCCAACUUGUUCUUCUC .-.---.(--------(((.....))))........(((((......))))).....(((((((.((((((..(((..((.....))..))).)))))).)..))))))........... ( -31.40) >DroAna_CAF1 9999 119 + 1 A-AGAUAUAUCUCCUAUACAUACUGUGUUCUAUUUCAAGGCCAAAGAGCUUGAUUUUGUUCGCCUCGUGCUGGGCUUUCUUCUUCAAGCGGCAGGCGCGGGAGGCCAGCUUGUUCUUCUC (-(((..........((((.....))))..........((((...((((........))))..((((((((..(((..(((....))).))).)))))))).)))).......))))... ( -29.90) >consensus C_AAAUUG________AACUCACUGUGCUCAAUUUCCAGGCCAUAUAGCUUGAUCUUGUUGGCCUCGUGCUGGGCCUUCUUCUUCAGGCGACAGGCGCGCGAGGCCAGCUUGUUCUUCUC .........................................................(((((((((.(((...((((((.((....)).)).))))))).)))))))))........... (-20.55 = -20.22 + -0.33)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:49:53 2006