
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 2,665,111 – 2,665,213 |
| Length | 102 |
| Max. P | 0.717318 |

| Location | 2,665,111 – 2,665,213 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 96.99 |
| Mean single sequence MFE | -21.72 |
| Consensus MFE | -20.44 |
| Energy contribution | -20.67 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -0.96 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.717318 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
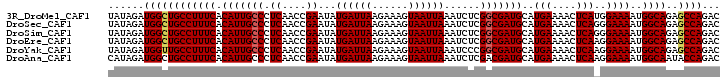
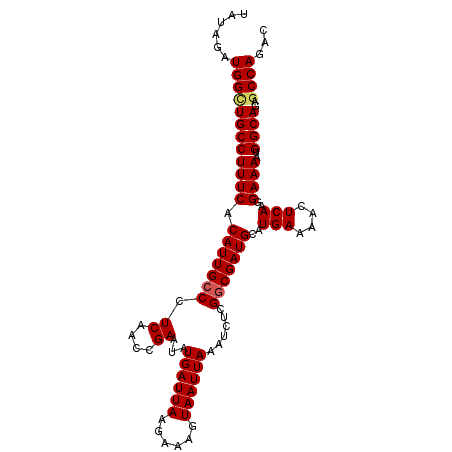
>3R_DroMel_CAF1 2665111 102 + 27905053 UAUAGAUGGCUGCCUUUCACAUUGCCCUCAACCGAAUAUGAUUAAGAAAGUAAUUAAAUCUCGGCGAUGCAUGAAAACUCAUGGAAAAUGGCAGAGCCAGAC ......((((((((((((.(((((((.((....))...((((((......))))))......)))))))(((((....)))))))))..)))..)))))... ( -26.80) >DroSec_CAF1 24608 102 + 1 UAUAGAUGGCUGCCUUUCACAUUGCCCUCAACCGAAUAUGAUUAAGAAAGUAAUUAAAUCUCGGCGAUGCAUGAAAACUCAGGGAAAAUGGCAGAGCCAGAC ......((((((((((((.(((((((.((....))...((((((......))))))......)))))))..(((....)))..))))..)))..)))))... ( -22.70) >DroSim_CAF1 17691 102 + 1 UAUAGAUGGCUGCCUUUCACAUUGCCCUCAACCGAAUAUGAUUAAGAAAGUAAUUAAAUCUCGGCGAUGCAUGAAAACUCAGGGAAAAUGGCAGAGCCAGAC ......((((((((((((.(((((((.((....))...((((((......))))))......)))))))..(((....)))..))))..)))..)))))... ( -22.70) >DroEre_CAF1 17636 102 + 1 UAUAGAUGGCUGCCUUUCACAUUGCCCUCAACCGAAUAUGAUUAAGAAAGUAAUUAAAUCUCGGCGAUGCAUGAAAACUCAAGGAAAAUGGCAGAGCCAGAC ......((((((((((((.(((((((.((....))...((((((......))))))......)))))))..(((....)))..))))..)))..)))))... ( -22.70) >DroYak_CAF1 24808 102 + 1 UAUAGAUGGUUGCCUUUCACAUUGCCCUCAACCGAAUAUGAUUAAGAAAGUAAUUAAAUCCCGGCGAUGCAUGAAAACUCAAGGAAAAUGGCAGAGCCAGAC ......((((((((((((.(((((((.((....))...((((((......))))))......)))))))..(((....)))..))))..))))..))))... ( -21.30) >DroAna_CAF1 12747 102 + 1 CAUAGAUGGCUGCCUUUCACAUUGCCCUCAACCGAAUAUGAUUAAGAAAGUAAUUAAAUCUCGACGAUGCAUGAAAACUCAAGGAAAAUGGCAAUACCAGAC ......(((.((((((((....(((..((...(((((.((((((......)))))).)).)))..)).)))(((....)))..))))..))))...)))... ( -14.10) >consensus UAUAGAUGGCUGCCUUUCACAUUGCCCUCAACCGAAUAUGAUUAAGAAAGUAAUUAAAUCUCGGCGAUGCAUGAAAACUCAAGGAAAAUGGCAGAGCCAGAC ......((((((((((((.(((((((.((....))...((((((......))))))......)))))))..(((....)))..))))..))))..))))... (-20.44 = -20.67 + 0.22)
| Location | 2,665,111 – 2,665,213 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 96.99 |
| Mean single sequence MFE | -23.58 |
| Consensus MFE | -22.45 |
| Energy contribution | -22.23 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.09 |
| Mean z-score | -0.80 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.573405 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
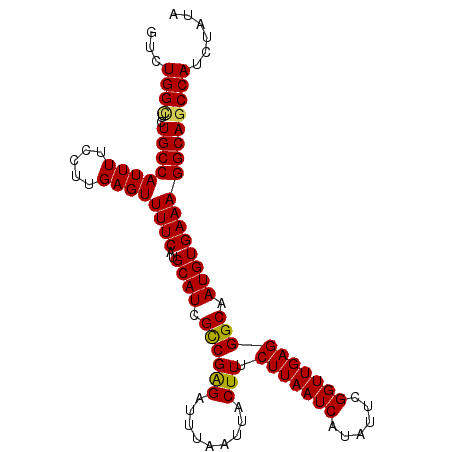
>3R_DroMel_CAF1 2665111 102 - 27905053 GUCUGGCUCUGCCAUUUUCCAUGAGUUUUCAUGCAUCGCCGAGAUUUAAUUACUUUCUUAAUCAUAUUCGGUUGAGGGCAAUGUGAAAGGCAGCCAUCUAUA ...(((((..(((.(((..(((..(((((((......((((((.....((((......))))....))))))))))))).)))..)))))))))))...... ( -25.10) >DroSec_CAF1 24608 102 - 1 GUCUGGCUCUGCCAUUUUCCCUGAGUUUUCAUGCAUCGCCGAGAUUUAAUUACUUUCUUAAUCAUAUUCGGUUGAGGGCAAUGUGAAAGGCAGCCAUCUAUA ...(((((..(((((((.....))))((((..((((.((((((.........))).(((((((......)))))))))).))))))))))))))))...... ( -23.70) >DroSim_CAF1 17691 102 - 1 GUCUGGCUCUGCCAUUUUCCCUGAGUUUUCAUGCAUCGCCGAGAUUUAAUUACUUUCUUAAUCAUAUUCGGUUGAGGGCAAUGUGAAAGGCAGCCAUCUAUA ...(((((..(((((((.....))))((((..((((.((((((.........))).(((((((......)))))))))).))))))))))))))))...... ( -23.70) >DroEre_CAF1 17636 102 - 1 GUCUGGCUCUGCCAUUUUCCUUGAGUUUUCAUGCAUCGCCGAGAUUUAAUUACUUUCUUAAUCAUAUUCGGUUGAGGGCAAUGUGAAAGGCAGCCAUCUAUA ...(((((..(((((((.....))))((((..((((.((((((.........))).(((((((......)))))))))).))))))))))))))))...... ( -23.70) >DroYak_CAF1 24808 102 - 1 GUCUGGCUCUGCCAUUUUCCUUGAGUUUUCAUGCAUCGCCGGGAUUUAAUUACUUUCUUAAUCAUAUUCGGUUGAGGGCAAUGUGAAAGGCAACCAUCUAUA ((((.((..((((....(((((((((......)).)))..))))............(((((((......)))))))))))..))...))))........... ( -22.20) >DroAna_CAF1 12747 102 - 1 GUCUGGUAUUGCCAUUUUCCUUGAGUUUUCAUGCAUCGUCGAGAUUUAAUUACUUUCUUAAUCAUAUUCGGUUGAGGGCAAUGUGAAAGGCAGCCAUCUAUG ((((..(((((((.......(((((((((.(((...))).))))))))).......(((((((......))))))))))))))....))))........... ( -23.10) >consensus GUCUGGCUCUGCCAUUUUCCUUGAGUUUUCAUGCAUCGCCGAGAUUUAAUUACUUUCUUAAUCAUAUUCGGUUGAGGGCAAUGUGAAAGGCAGCCAUCUAUA ...((((..((((((((.....))))((((..((((.((((((.........))).(((((((......)))))))))).))))))))))))))))...... (-22.45 = -22.23 + -0.22)

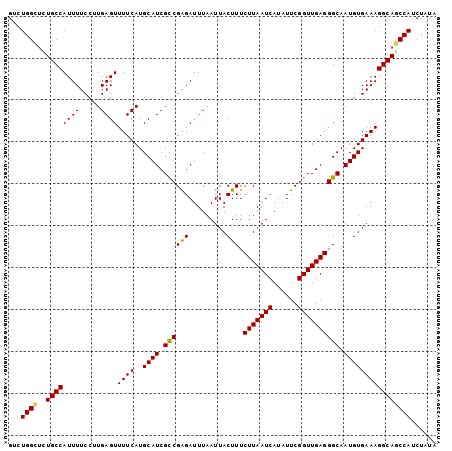
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:51:27 2006