| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 20,349,435 – 20,349,560 |
| Length | 125 |
| Max. P | 0.991005 |

| Location | 20,349,435 – 20,349,530 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 76.78 |
| Mean single sequence MFE | -26.15 |
| Consensus MFE | -12.59 |
| Energy contribution | -13.01 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.48 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.914182 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20349435 95 + 27905053 AUUUUUGGCCAAACACUUGCUG-------GCAGACUCAUGU----CCACAC--GCAUACAAGUGGAUGCCUAAGCCCAGUUGGACAAAAAAAUGUGCUGGCCAAACGA ...((((((((.(((...((((-------((((((....))----)..(((--........)))..))))..)))((....)).........)))..))))))))... ( -27.50) >DroSec_CAF1 157986 95 + 1 AUUUUUGGCCAAACACUUGCUA-------GCAGACUCAUGU----CCACAC--GCACACAAGUGCAUGCCUAAGCCCAGUUGGACAAAAGAAUGUGCUGGCCAAACGA ...((((((((.(((.(((...-------.))).((..(((----(((.((--((((....))))..((....))...))))))))..))..)))..))))))))... ( -29.30) >DroSim_CAF1 167152 95 + 1 AUUUUUGGCCAAACACUUGCCA-------GCAGACUCAUGU----CCACAC--GCACACAAGUGCAUGCCUAAGCCCAGUUGGACAAAAGAAUGUGCUGGCCAAACGA ...((((((((.((((((.(((-------((.(((....))----).....--((((....)))).............)))))....)))..)))..))))))))... ( -29.70) >DroEre_CAF1 163805 95 + 1 AUUUUUGGCCAAACACUUGCUC-------GCAGACUCAUGU----UCAAAC--GCACACAAGUGCAUGCCUAAGCCCAGUUGGACAAAAGAAUGUGCUGGCCAAACGA ...((((((((.(((.(((...-------.))).((..(((----((((..--((((....))))..((....))....)))))))..))..)))..))))))))... ( -26.60) >DroYak_CAF1 166519 73 + 1 AUUUUUGGCCAAACACUUGCUA-------GCAGACUCA----------------------------UGCCUAAGCGCAGUUGGACAAAAGAAUGUGCUGGCCAAACGA ...(((((((............-------(((......----------------------------)))...((((((.((........)).)))))))))))))... ( -20.20) >DroAna_CAF1 164697 108 + 1 UUUUUUGGCUAAGCACUCUUAGACACAUAAAAGACGGAUUUACACUCACACUUACACACAACUGGCAGUAAUCGGCCAGUUAGAAAAAAGAAUGUGCUGUCCGAACGA ((((.((.(((((....)))))...)).))))..(((((...(((.....(((......(((((((........)))))))......)))...)))..)))))..... ( -23.60) >consensus AUUUUUGGCCAAACACUUGCUA_______GCAGACUCAUGU____CCACAC__GCACACAAGUGCAUGCCUAAGCCCAGUUGGACAAAAGAAUGUGCUGGCCAAACGA ...((((((((.....((((.........))))....................(((((..........((...........)).........)))))))))))))... (-12.59 = -13.01 + 0.42)

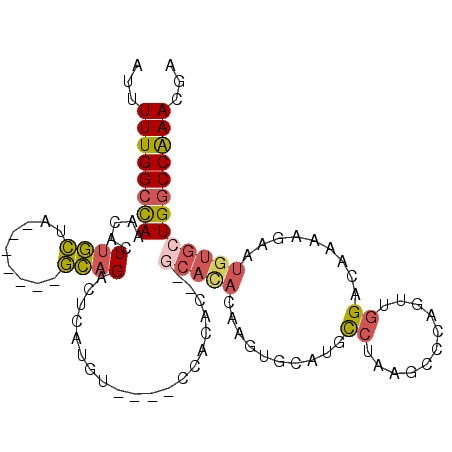

| Location | 20,349,435 – 20,349,530 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 76.78 |
| Mean single sequence MFE | -32.33 |
| Consensus MFE | -17.91 |
| Energy contribution | -18.55 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.85 |
| Structure conservation index | 0.55 |
| SVM decision value | 2.24 |
| SVM RNA-class probability | 0.991005 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20349435 95 - 27905053 UCGUUUGGCCAGCACAUUUUUUUGUCCAACUGGGCUUAGGCAUCCACUUGUAUGC--GUGUGG----ACAUGAGUCUGC-------CAGCAAGUGUUUGGCCAAAAAU ...((((((((((((........((((....))))...(((((((((.((....)--).))))----)........)))-------).....))).)))))))))... ( -34.00) >DroSec_CAF1 157986 95 - 1 UCGUUUGGCCAGCACAUUCUUUUGUCCAACUGGGCUUAGGCAUGCACUUGUGUGC--GUGUGG----ACAUGAGUCUGC-------UAGCAAGUGUUUGGCCAAAAAU ...((((((((((((...((...((((....))))..))((((((((....))))--))(..(----((....)))..)-------..))..))).)))))))))... ( -38.70) >DroSim_CAF1 167152 95 - 1 UCGUUUGGCCAGCACAUUCUUUUGUCCAACUGGGCUUAGGCAUGCACUUGUGUGC--GUGUGG----ACAUGAGUCUGC-------UGGCAAGUGUUUGGCCAAAAAU ...((((((((((((..((...(((((..(((....)))((((((((....))))--))))))----))).))(((...-------.)))..))).)))))))))... ( -39.10) >DroEre_CAF1 163805 95 - 1 UCGUUUGGCCAGCACAUUCUUUUGUCCAACUGGGCUUAGGCAUGCACUUGUGUGC--GUUUGA----ACAUGAGUCUGC-------GAGCAAGUGUUUGGCCAAAAAU ...((((((((((((...((...((((....))))..))((.((((((..(((..--......----)))..)))..))-------).))..))).)))))))))... ( -35.20) >DroYak_CAF1 166519 73 - 1 UCGUUUGGCCAGCACAUUCUUUUGUCCAACUGCGCUUAGGCA----------------------------UGAGUCUGC-------UAGCAAGUGUUUGGCCAAAAAU ...((((((((((((........((......))((((((((.----------------------------...))))).-------.)))..))).)))))))))... ( -22.90) >DroAna_CAF1 164697 108 - 1 UCGUUCGGACAGCACAUUCUUUUUUCUAACUGGCCGAUUACUGCCAGUUGUGUGUAAGUGUGAGUGUAAAUCCGUCUUUUAUGUGUCUAAGAGUGCUUAGCCAAAAAA .....((((....((((((.(((..(((((((((........)))))))).)...)))...))))))...)))).........((.(((((....))))).))..... ( -24.10) >consensus UCGUUUGGCCAGCACAUUCUUUUGUCCAACUGGGCUUAGGCAUGCACUUGUGUGC__GUGUGG____ACAUGAGUCUGC_______UAGCAAGUGUUUGGCCAAAAAU ...((((((((((((........((((....)))).(((((................................)))))..............)))).))))))))... (-17.91 = -18.55 + 0.64)


| Location | 20,349,457 – 20,349,560 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 81.20 |
| Mean single sequence MFE | -25.83 |
| Consensus MFE | -15.31 |
| Energy contribution | -17.37 |
| Covariance contribution | 2.06 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.677794 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20349457 103 + 27905053 -GCAGACUCAUGU----CCACAC--GCAUACAAGUGGAUGCCUAAGCCCAGUUGGACAAAAAAAUGUGCUGGCCAAACGAAAGACUUAGGCAACAAACUUGAAUUCGGUA -...(((....))----).((.(--(.((.(((((...((((((((.(((((...(((......)))))))).....(....).))))))))....))))).)).)))). ( -28.20) >DroSec_CAF1 158008 103 + 1 -GCAGACUCAUGU----CCACAC--GCACACAAGUGCAUGCCUAAGCCCAGUUGGACAAAAGAAUGUGCUGGCCAAACGAAAGACUUAGGUAACAAACUUGAAUUCGGUA -(((.(.((.(((----(((.((--((((....))))..((....))...))))))))...)).).)))..(((...(....)..((((((.....))))))....))). ( -25.80) >DroSim_CAF1 167174 103 + 1 -GCAGACUCAUGU----CCACAC--GCACACAAGUGCAUGCCUAAGCCCAGUUGGACAAAAGAAUGUGCUGGCCAAACGAAAGACUUAGGCAACAAACUUGAAUUCGGCA -((.((.(((.((----......--((((....)))).((((((((.(((((...(((......)))))))).....(....).))))))))....)).)))..)).)). ( -28.90) >DroEre_CAF1 163827 103 + 1 -GCAGACUCAUGU----UCAAAC--GCACACAAGUGCAUGCCUAAGCCCAGUUGGACAAAAGAAUGUGCUGGCCAAACGAAAGACUUAGGCAACAAACUUGAAUUCGGCA -((.((.....((----((((..--((((....)))).((((((((.(((((...(((......)))))))).....(....).))))))))......)))))))).)). ( -30.20) >DroYak_CAF1 166541 81 + 1 -GCAGACUCA----------------------------UGCCUAAGCGCAGUUGGACAAAAGAAUGUGCUGGCCAAACGAAAGACUUAGGCAACAAACUUGAAUUCGGCA -((.((.(((----------------------------((((((((....((..((((......))).)..))....(....).)))))))).......)))..)).)). ( -19.81) >DroAna_CAF1 164725 107 + 1 AAAAGACGGAUUUACACUCACACUUACACACAACUGGCAGUAAUCGGCCAGUUAGAAAAAAGAAUGUGCUGUCCGAACGAAAGACUUAGGCAACAAACUUGAAUUCA--- ..(((.(((((...(((.....(((......(((((((........)))))))......)))...)))..)))))..(....).))).(....).............--- ( -22.10) >consensus _GCAGACUCAUGU____CCACAC__GCACACAAGUGCAUGCCUAAGCCCAGUUGGACAAAAGAAUGUGCUGGCCAAACGAAAGACUUAGGCAACAAACUUGAAUUCGGCA ..............................(((((...((((((((.(((((...(((......)))))))).....(....).))))))))....)))))......... (-15.31 = -17.37 + 2.06)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:49:39 2006