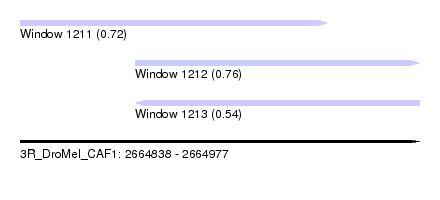
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 2,664,838 – 2,664,977 |
| Length | 139 |
| Max. P | 0.756584 |

| Location | 2,664,838 – 2,664,945 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 81.99 |
| Mean single sequence MFE | -28.09 |
| Consensus MFE | -17.65 |
| Energy contribution | -18.73 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.723156 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2664838 107 + 27905053 AAAGAGGCUAGCCUAAUUGUCCAGCAGGCUGUUGACUUUUGUGUUCCUAAA-UAAUAGGGCAAUUAAGUGCAGGGUUACCAAAAGCUAUAACUGCACAUGGUUCUAUA ..((((.(((...((((((((((((.....)))).((.((((........)-))).)))))))))).((((((((((......))))....)))))).)))))))... ( -24.60) >DroSec_CAF1 24335 107 + 1 AAAGAGCCUAGCCUAAUUGUCCACCUGGCUGUUUACUUUUGUGUUCCUAAA-GAAGAGGGCAAUUAAAUGCAGGGCUACCAAAAGGAAUAACUGCACAUGGUUCUAUA ..((((((.....(((((((((..((....(..(((....)))..)....)-)....)))))))))..(((((.....((....)).....)))))...))))))... ( -26.60) >DroSim_CAF1 17429 107 + 1 AAAGAGCCUAGCCUAAUUGUCCACCAGGCUGUUGACUUUUGUGUUCCUAAA-GAAGAGGGCAAUUAAGUGCAGGGUUACCGAAAGGUAUAACUGCACAUAGUUCUAUA ..(((((......(((((((((..(((....))).(((((..........)-)))).))))))))).((((((...((((....))))...))))))...)))))... ( -35.30) >DroEre_CAF1 17376 107 + 1 AAAGUGGCUAGCCUAAUUGUCCACCAGGCUGUUGACUUUUGUGUUCGUAAAAAAAGAGGUCAAUUAAGUGCAGCGUUACCAA-AGGUAUAGCUGCACAUAGUUCUUUA ((((.(((((((((...........)))).((((((((((..............))))))))))...(((((((..((((..-.))))..))))))).))))))))). ( -32.14) >DroYak_CAF1 24581 86 + 1 AAAGUGGCUAGGCUAAUUGUCCACCAGGCUG------------UUCCUAAAAAAAGAGGUCAAUU-AGUGCAGGGUUGCCAAAAGGUACAACUGCACAA--------- ..((..(((.((...........)).)))..------------))(((........)))......-.((((((...((((....))))...))))))..--------- ( -21.80) >consensus AAAGAGGCUAGCCUAAUUGUCCACCAGGCUGUUGACUUUUGUGUUCCUAAA_AAAGAGGGCAAUUAAGUGCAGGGUUACCAAAAGGUAUAACUGCACAUAGUUCUAUA .............(((((((((...................................))))))))).((((((...((((....))))...))))))........... (-17.65 = -18.73 + 1.08)

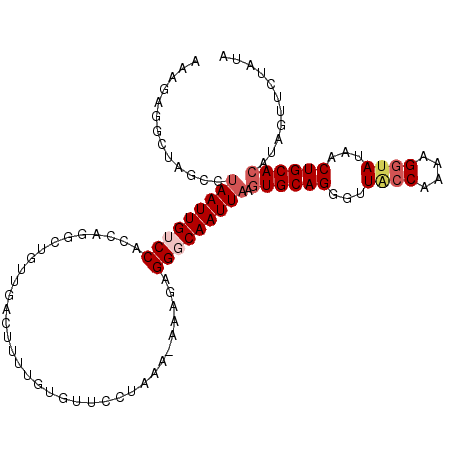
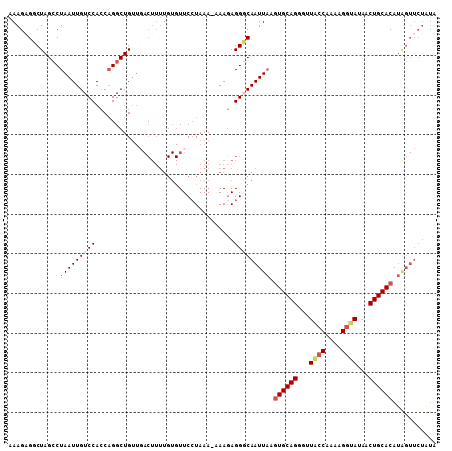
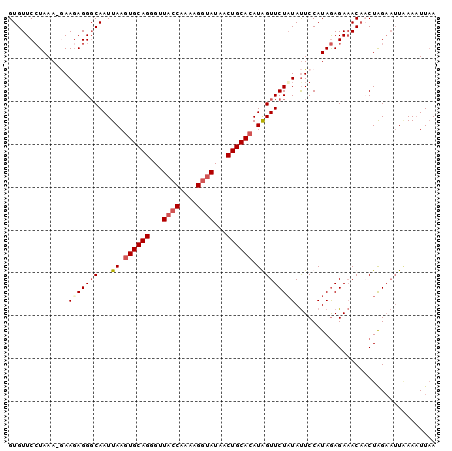
| Location | 2,664,878 – 2,664,977 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 85.43 |
| Mean single sequence MFE | -23.07 |
| Consensus MFE | -13.88 |
| Energy contribution | -15.00 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.756584 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
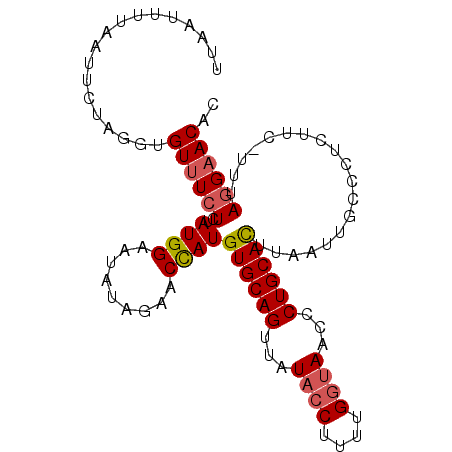
>3R_DroMel_CAF1 2664878 99 + 27905053 GUGUUCCUAAA-UAAUAGGGCAAUUAAGUGCAGGGUUACCAAAAGCUAUAACUGCACAUGGUUCUAUAUUCCAUAGAGAAACACCUAGAAUUGAAAUUAA (((((((((..-.((((.((.(((((.((((((((((......))))....)))))).))))))).))))...))).).)))))................ ( -19.40) >DroSec_CAF1 24375 99 + 1 GUGUUCCUAAA-GAAGAGGGCAAUUAAAUGCAGGGCUACCAAAAGGAAUAACUGCACAUGGUUCUAUAUUCCAUAGAGAAACAACUAGAAUUAAAAUUUA ((.((((((..-(((.(((((.......(((((.....((....)).....)))))....)))))...)))..))).)))...))............... ( -17.60) >DroSim_CAF1 17469 99 + 1 GUGUUCCUAAA-GAAGAGGGCAAUUAAGUGCAGGGUUACCGAAAGGUAUAACUGCACAUAGUUCUAUAUUCCAUAGAGAAACAACUAGAAUUAAAUUUAA ((.((((((..-(((...((.(((((.((((((...((((....))))...)))))).)))))))...)))..))).)))...))............... ( -29.10) >DroEre_CAF1 17416 99 + 1 GUGUUCGUAAAAAAAGAGGUCAAUUAAGUGCAGCGUUACCAA-AGGUAUAGCUGCACAUAGUUCUUUACUCCAUAAAGAUACCCCUGGCCAUUAAAUUAA .................(((((((((.(((((((..((((..-.))))..))))))).))))((((((.....))))))......))))).......... ( -26.20) >consensus GUGUUCCUAAA_GAAGAGGGCAAUUAAGUGCAGGGUUACCAAAAGGUAUAACUGCACAUAGUUCUAUAUUCCAUAGAGAAACAACUAGAAUUAAAAUUAA ..............(((((((...((.((((((...((((....))))...)))))).)))))))))................................. (-13.88 = -15.00 + 1.12)

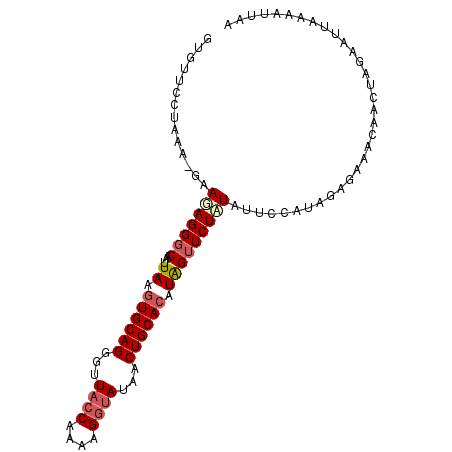
| Location | 2,664,878 – 2,664,977 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 100 |
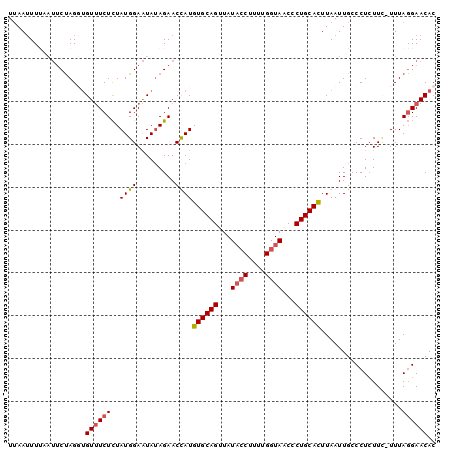
| Reading direction | reverse |
| Mean pairwise identity | 85.43 |
| Mean single sequence MFE | -20.10 |
| Consensus MFE | -14.26 |
| Energy contribution | -14.83 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.544619 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2664878 99 - 27905053 UUAAUUUCAAUUCUAGGUGUUUCUCUAUGGAAUAUAGAACCAUGUGCAGUUAUAGCUUUUGGUAACCCUGCACUUAAUUGCCCUAUUA-UUUAGGAACAC ..........((((..(((((((.....)))))))))))....((((((...((.(....).))...))))))........((((...-..))))..... ( -18.50) >DroSec_CAF1 24375 99 - 1 UAAAUUUUAAUUCUAGUUGUUUCUCUAUGGAAUAUAGAACCAUGUGCAGUUAUUCCUUUUGGUAGCCCUGCAUUUAAUUGCCCUCUUC-UUUAGGAACAC .................(((((((....((((...........((((((.....((....)).....))))))............)))-)..))))))). ( -16.40) >DroSim_CAF1 17469 99 - 1 UUAAAUUUAAUUCUAGUUGUUUCUCUAUGGAAUAUAGAACUAUGUGCAGUUAUACCUUUCGGUAACCCUGCACUUAAUUGCCCUCUUC-UUUAGGAACAC .................((((((((((((...)))))).....((((((...((((....))))...))))))...............-....)))))). ( -20.10) >DroEre_CAF1 17416 99 - 1 UUAAUUUAAUGGCCAGGGGUAUCUUUAUGGAGUAAAGAACUAUGUGCAGCUAUACCU-UUGGUAACGCUGCACUUAAUUGACCUCUUUUUUUACGAACAC ..............((((((.(((((((...))))))).....(((((((..((((.-..))))..))))))).......)))))).............. ( -25.40) >consensus UUAAUUUUAAUUCUAGGUGUUUCUCUAUGGAAUAUAGAACCAUGUGCAGUUAUACCUUUUGGUAACCCUGCACUUAAUUGCCCUCUUC_UUUAGGAACAC ..................((((((..((((.........))))((((((...((((....))))...))))))...................)))))).. (-14.26 = -14.83 + 0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:51:26 2006