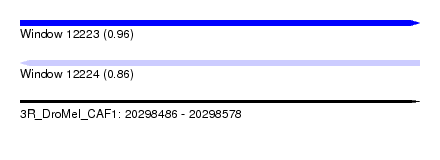
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 20,298,486 – 20,298,578 |
| Length | 92 |
| Max. P | 0.964920 |

| Location | 20,298,486 – 20,298,578 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 77.79 |
| Mean single sequence MFE | -26.98 |
| Consensus MFE | -20.51 |
| Energy contribution | -20.60 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.964920 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
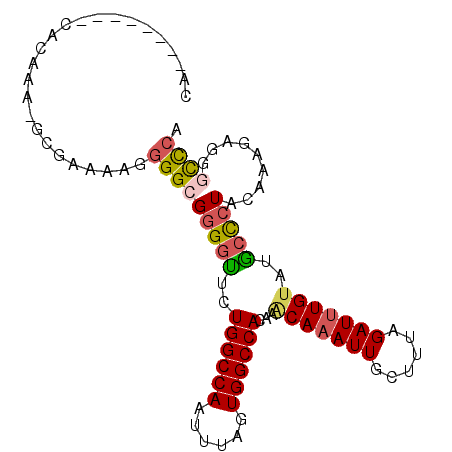
>3R_DroMel_CAF1 20298486 92 + 27905053 -----AGAGGCACAUA-GUGAAAAGGGGCGGGGUUCUGGCCAAUUUAGUGGCCACAAACAAAUUGCUUUAGAUUUGUUUGCCCUACAAAGAGAGUCCA -----...........-........((((((((...((((((......))))))((((((((((......)))))))))))))).(.....).)))). ( -27.10) >DroSec_CAF1 107344 95 + 1 CA--GAGAGACGUACA-GUGAAAAGGGGCGGGGUUCUGGCCAAUUUAGUGGCCACAAACAAAUUGCUUUAGAUUUGUAUGCCCUACAAAGAAGGCCCA ..--............-........(((((((((..((((((......))))))...(((((((......)))))))..))))).(......))))). ( -27.20) >DroSim_CAF1 115683 97 + 1 CAUACAGCGACACAGA-GCGAAAAGGGGCGGGGUUCUGGCCAAUUUAGUGGCCACAAACAAAUUGCUUUAGAUUUGUAUGCCCUACAAAGAGGGCCCA .(((((((........-)).....(((((((.(((.((((((......))))))..)))...))))))).....)))))(((((......)))))... ( -29.30) >DroEre_CAF1 110375 90 + 1 CA--------UACAAAAGGAAAAAGGGGCGGGGCAAUGGCCAAUUUAGUGGCCACAAGCAAAUUGCUUUAGAUUUGUAUGCCCUACAAAGUGGACUCA ..--------.................(..(((((.((((((......))))))...(((((((......))))))).)))))..)............ ( -25.50) >DroYak_CAF1 112864 90 + 1 CA--------CACAAAGGGAAAAGAGGGCGGGGCUAUGGCCAAUUUAGUGGCCACAAACAAAUUGCUUUAGAUUUGUAUGCCCUACAAAGUGGGCUCA ..--------...............(((((((((..((((((......))))))...(((((((......)))))))..))))).(.....).)))). ( -27.00) >DroPer_CAF1 130024 87 + 1 CA--------CACACA-GCGACAG--GGAGUGUUGUUGGCCAAUUUAGUGGCCACAAACAAAUUGCUCUAGAUCUGAGCACUCUUCCCAAUAGAUUCA ..--------......-......(--((((((((..((((((......))))))..))))...(((((.......)))))...))))).......... ( -25.80) >consensus CA________CACAAA_GCGAAAAGGGGCGGGGUUCUGGCCAAUUUAGUGGCCACAAACAAAUUGCUUUAGAUUUGUAUGCCCUACAAAGAGGGCCCA .........................(((((((((..((((((......))))))...(((((((......)))))))..))))).........)))). (-20.51 = -20.60 + 0.09)

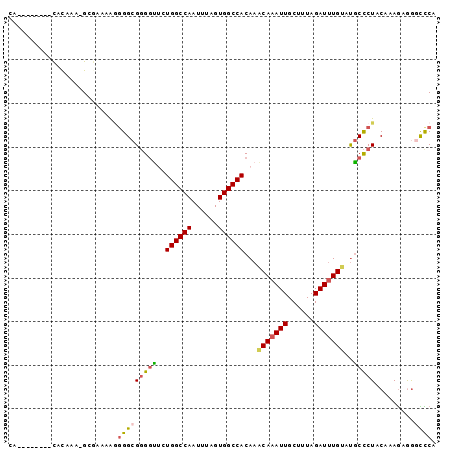
| Location | 20,298,486 – 20,298,578 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 77.79 |
| Mean single sequence MFE | -23.77 |
| Consensus MFE | -16.51 |
| Energy contribution | -16.73 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.855449 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
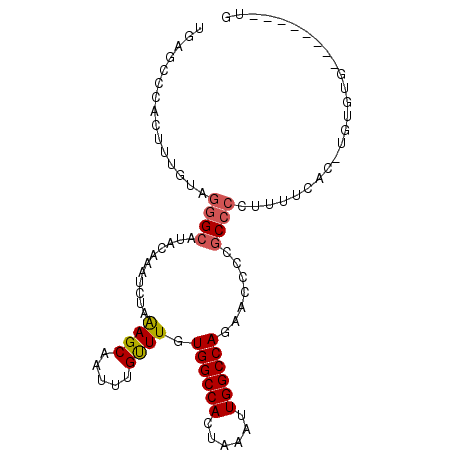
>3R_DroMel_CAF1 20298486 92 - 27905053 UGGACUCUCUUUGUAGGGCAAACAAAUCUAAAGCAAUUUGUUUGUGGCCACUAAAUUGGCCAGAACCCCGCCCCUUUUCAC-UAUGUGCCUCU----- .((((.........(((((((((((((........)))))))))((((((......)))))).........))))......-...)).))...----- ( -23.67) >DroSec_CAF1 107344 95 - 1 UGGGCCUUCUUUGUAGGGCAUACAAAUCUAAAGCAAUUUGUUUGUGGCCACUAAAUUGGCCAGAACCCCGCCCCUUUUCAC-UGUACGUCUCUC--UG .((((...(..((..((((..((((((........))))))((.((((((......)))))).))....)))).....)).-.)...))))...--.. ( -22.30) >DroSim_CAF1 115683 97 - 1 UGGGCCCUCUUUGUAGGGCAUACAAAUCUAAAGCAAUUUGUUUGUGGCCACUAAAUUGGCCAGAACCCCGCCCCUUUUCGC-UCUGUGUCGCUGUAUG ...(((((......)))))(((((........((.....((((.((((((......))))))))))...))........((-........))))))). ( -23.40) >DroEre_CAF1 110375 90 - 1 UGAGUCCACUUUGUAGGGCAUACAAAUCUAAAGCAAUUUGCUUGUGGCCACUAAAUUGGCCAUUGCCCCGCCCCUUUUUCCUUUUGUA--------UG ............(..(((((..........((((.....))))(((((((......))))))))))))..).................--------.. ( -23.80) >DroYak_CAF1 112864 90 - 1 UGAGCCCACUUUGUAGGGCAUACAAAUCUAAAGCAAUUUGUUUGUGGCCACUAAAUUGGCCAUAGCCCCGCCCUCUUUUCCCUUUGUG--------UG ...((((........))))(((((((...((((........(((((((((......))))))))).........))))....))))))--------). ( -25.03) >DroPer_CAF1 130024 87 - 1 UGAAUCUAUUGGGAAGAGUGCUCAGAUCUAGAGCAAUUUGUUUGUGGCCACUAAAUUGGCCAACAACACUCC--CUGUCGC-UGUGUG--------UG ..........((((....(((((.......)))))...((((..((((((......))))))..)))).)))--)...(((-.....)--------)) ( -24.40) >consensus UGAGCCCACUUUGUAGGGCAUACAAAUCUAAAGCAAUUUGUUUGUGGCCACUAAAUUGGCCAGAACCCCGCCCCUUUUCAC_UGUGUG________UG ...............((((...........((((.....)))).((((((......)))))).......))))......................... (-16.51 = -16.73 + 0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:49:13 2006