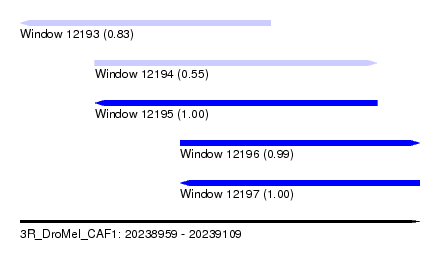
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 20,238,959 – 20,239,109 |
| Length | 150 |
| Max. P | 0.999968 |

| Location | 20,238,959 – 20,239,053 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 94.71 |
| Mean single sequence MFE | -20.86 |
| Consensus MFE | -20.80 |
| Energy contribution | -20.80 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.15 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.834121 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20238959 94 - 27905053 UGCUCCUUUGACCACCAUGAAACCCCAACCCAAUAUCAACUCUGGUAAUCCCCAGUGUGAUGGCUCCCACUUGGCGUGGUCGUAUAAAUAUAUA-- .........((((((..................(((((...((((......))))..)))))(((.......))))))))).............-- ( -20.80) >DroSec_CAF1 50038 94 - 1 UGCUCCGUUGACCACCAUGAAACCCCAACCCAAUAUCAACUCUGGUAAUCCCCAGUGUGAUGGCUCCCACUUGGCGUGGUCGUAUAAAUAUAUA-- .........((((((..................(((((...((((......))))..)))))(((.......))))))))).............-- ( -20.80) >DroSim_CAF1 54027 92 - 1 UGCUCCGUUGACCACCAUGAAACCCCAACCCGAUAUCAACUCUGGUAAUCCCCAGUGUGAUGGCUCCCACUUGGCGUGGUCGUAUAAACAUA---- ......(((.((.((((((.....((((.....(((((...((((......))))..)))))........)))))))))).))...)))...---- ( -20.92) >DroEre_CAF1 51196 96 - 1 CGCUCCGUUGACCACCAUGAAACCCCAACCCAAUAUCAACUCUGGUAAUCCCCAGUGUGAUGGCUCCCACUUGGCGUGGUCGUAUAAAUACAUAUA .........((((((..................(((((...((((......))))..)))))(((.......)))))))))(((....)))..... ( -21.00) >DroYak_CAF1 52398 91 - 1 UGCUCCGUUGACCACCGUGAAACCCCAACCCAAUAUCAACUCUGGUAAUCCCCAGUGUGAUGGCUCCCACUUGGCGUGGUCGUAUAAAUAC----- .........((((((..................(((((...((((......))))..)))))(((.......)))))))))..........----- ( -20.80) >consensus UGCUCCGUUGACCACCAUGAAACCCCAACCCAAUAUCAACUCUGGUAAUCCCCAGUGUGAUGGCUCCCACUUGGCGUGGUCGUAUAAAUAUAUA__ .........((((((..................(((((...((((......))))..)))))(((.......)))))))))............... (-20.80 = -20.80 + 0.00)


| Location | 20,238,987 – 20,239,093 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 97.74 |
| Mean single sequence MFE | -36.74 |
| Consensus MFE | -35.34 |
| Energy contribution | -35.38 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.13 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.553692 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20238987 106 + 27905053 GAGCCAUCACACUGGGGAUUACCAGAGUUGAUAUUGGGUUGGGGUUUCAUGGUGGUCAAAGGAGCAGGAGCACGCUCGCUUUCGUGCUGACCAAGCACGAAAUUCC .(((((((((.((((......)))).).))))....))))((((((((.((.((((((...((((........))))((......))))))))..)).)))))))) ( -36.20) >DroSec_CAF1 50066 106 + 1 GAGCCAUCACACUGGGGAUUACCAGAGUUGAUAUUGGGUUGGGGUUUCAUGGUGGUCAACGGAGCAGGAGCACGCUCGCUUUCGUGCUGACCAAGCACGAAAUUCC .(((((((((.((((......)))).).))))....))))((((((((.((.(((((((((((((..(((....))))).)))))..))))))..)).)))))))) ( -37.20) >DroSim_CAF1 54053 106 + 1 GAGCCAUCACACUGGGGAUUACCAGAGUUGAUAUCGGGUUGGGGUUUCAUGGUGGUCAACGGAGCAGGAGCACGCUCGCUUUCGUGCUGACCAAGCACGAAAUUCC .(((((((((.((((......)))).).))))....))))((((((((.((.(((((((((((((..(((....))))).)))))..))))))..)).)))))))) ( -37.20) >DroEre_CAF1 51226 106 + 1 GAGCCAUCACACUGGGGAUUACCAGAGUUGAUAUUGGGUUGGGGUUUCAUGGUGGUCAACGGAGCGGGAGCACGCUUGCUUUUGUGCUGACCAAGCACGAAAUUCC .(((((((((.((((......)))).).))))....))))((((((((.((.((((((.....(((((((((....)))))))))..))))))..)).)))))))) ( -37.80) >DroYak_CAF1 52423 106 + 1 GAGCCAUCACACUGGGGAUUACCAGAGUUGAUAUUGGGUUGGGGUUUCACGGUGGUCAACGGAGCAGGAGCACGCUCGCUUUCGUGCUGACCAAGCACGAAAUUCC .(((((((((.((((......)))).).))))....))))((((((((....(((((((((((((..(((....))))).)))))..)))))).....)))))))) ( -35.30) >consensus GAGCCAUCACACUGGGGAUUACCAGAGUUGAUAUUGGGUUGGGGUUUCAUGGUGGUCAACGGAGCAGGAGCACGCUCGCUUUCGUGCUGACCAAGCACGAAAUUCC .(((((((((.((((......)))).).))))....))))((((((((.((.((((((...((((........))))((......))))))))..)).)))))))) (-35.34 = -35.38 + 0.04)


| Location | 20,238,987 – 20,239,093 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 97.74 |
| Mean single sequence MFE | -34.46 |
| Consensus MFE | -34.04 |
| Energy contribution | -34.12 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.99 |
| SVM decision value | 3.78 |
| SVM RNA-class probability | 0.999606 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20238987 106 - 27905053 GGAAUUUCGUGCUUGGUCAGCACGAAAGCGAGCGUGCUCCUGCUCCUUUGACCACCAUGAAACCCCAACCCAAUAUCAACUCUGGUAAUCCCCAGUGUGAUGGCUC ((..(((((((..((((((((......))(((((......)))))...)))))).)))))))..)).......(((((...((((......))))..))))).... ( -33.30) >DroSec_CAF1 50066 106 - 1 GGAAUUUCGUGCUUGGUCAGCACGAAAGCGAGCGUGCUCCUGCUCCGUUGACCACCAUGAAACCCCAACCCAAUAUCAACUCUGGUAAUCCCCAGUGUGAUGGCUC ((..(((((((..((((((((.(....).(((((......))))).)))))))).)))))))..)).......(((((...((((......))))..))))).... ( -37.00) >DroSim_CAF1 54053 106 - 1 GGAAUUUCGUGCUUGGUCAGCACGAAAGCGAGCGUGCUCCUGCUCCGUUGACCACCAUGAAACCCCAACCCGAUAUCAACUCUGGUAAUCCCCAGUGUGAUGGCUC ((..(((((((..((((((((.(....).(((((......))))).)))))))).)))))))..)).......(((((...((((......))))..))))).... ( -37.00) >DroEre_CAF1 51226 106 - 1 GGAAUUUCGUGCUUGGUCAGCACAAAAGCAAGCGUGCUCCCGCUCCGUUGACCACCAUGAAACCCCAACCCAAUAUCAACUCUGGUAAUCCCCAGUGUGAUGGCUC ((..(((((((..((((((((.(....)..((((......))))..)))))))).)))))))..)).......(((((...((((......))))..))))).... ( -31.40) >DroYak_CAF1 52423 106 - 1 GGAAUUUCGUGCUUGGUCAGCACGAAAGCGAGCGUGCUCCUGCUCCGUUGACCACCGUGAAACCCCAACCCAAUAUCAACUCUGGUAAUCCCCAGUGUGAUGGCUC ((..(((((((..((((((((.(....).(((((......))))).)))))))).)))))))..)).......(((((...((((......))))..))))).... ( -33.60) >consensus GGAAUUUCGUGCUUGGUCAGCACGAAAGCGAGCGUGCUCCUGCUCCGUUGACCACCAUGAAACCCCAACCCAAUAUCAACUCUGGUAAUCCCCAGUGUGAUGGCUC ((..(((((((..((((((((.(....).(((((......))))).)))))))).)))))))..)).......(((((...((((......))))..))))).... (-34.04 = -34.12 + 0.08)

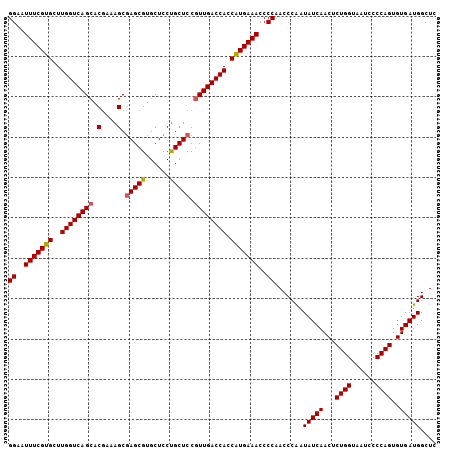
| Location | 20,239,019 – 20,239,109 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 97.33 |
| Mean single sequence MFE | -30.76 |
| Consensus MFE | -29.02 |
| Energy contribution | -28.90 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.46 |
| SVM RNA-class probability | 0.994221 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20239019 90 + 27905053 AUUGGGUUGGGGUUUCAUGGUGGUCAAAGGAGCAGGAGCACGCUCGCUUUCGUGCUGACCAAGCACGAAAUUCCCCGAUAAUUGCAAAUA .....(((((((((((.((.((((((...((((........))))((......))))))))..)).))))..)))))))........... ( -29.80) >DroSec_CAF1 50098 90 + 1 AUUGGGUUGGGGUUUCAUGGUGGUCAACGGAGCAGGAGCACGCUCGCUUUCGUGCUGACCAAGCACGAAAUUCCCCGAUAAUUGCAAAUA .....(((((((((((.((.(((((((((((((..(((....))))).)))))..))))))..)).))))..)))))))........... ( -30.80) >DroSim_CAF1 54085 90 + 1 AUCGGGUUGGGGUUUCAUGGUGGUCAACGGAGCAGGAGCACGCUCGCUUUCGUGCUGACCAAGCACGAAAUUCCCCGAUAAUUGCAAAUA ((((((...(((((((.((.(((((((((((((..(((....))))).)))))..))))))..)).)))))))))))))........... ( -32.40) >DroEre_CAF1 51258 90 + 1 AUUGGGUUGGGGUUUCAUGGUGGUCAACGGAGCGGGAGCACGCUUGCUUUUGUGCUGACCAAGCACGAAAUUCCCCGAUAAUUGCAAAUA .....(((((((((((.((.((((((.....(((((((((....)))))))))..))))))..)).))))..)))))))........... ( -31.40) >DroYak_CAF1 52455 90 + 1 AUUGGGUUGGGGUUUCACGGUGGUCAACGGAGCAGGAGCACGCUCGCUUUCGUGCUGACCAAGCACGAAAUUCCCCGAUAAUUGCAAAUA .....(((((((..(..((.(((((((((((((..(((....))))).)))))..))))))....))..)..)))))))........... ( -29.40) >consensus AUUGGGUUGGGGUUUCAUGGUGGUCAACGGAGCAGGAGCACGCUCGCUUUCGUGCUGACCAAGCACGAAAUUCCCCGAUAAUUGCAAAUA ((((((...(((((((.((.((((((...((((........))))((......))))))))..)).)))))))))))))........... (-29.02 = -28.90 + -0.12)


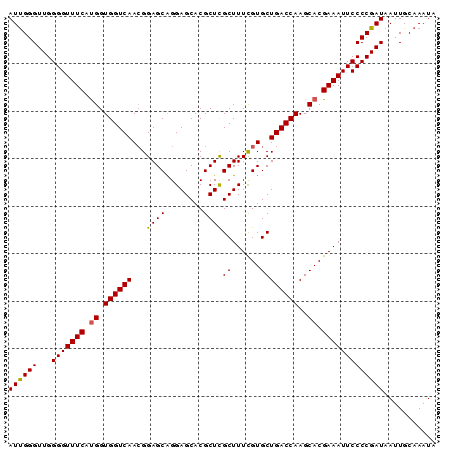
| Location | 20,239,019 – 20,239,109 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 97.33 |
| Mean single sequence MFE | -30.58 |
| Consensus MFE | -30.14 |
| Energy contribution | -30.22 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.10 |
| Structure conservation index | 0.99 |
| SVM decision value | 5.01 |
| SVM RNA-class probability | 0.999968 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20239019 90 - 27905053 UAUUUGCAAUUAUCGGGGAAUUUCGUGCUUGGUCAGCACGAAAGCGAGCGUGCUCCUGCUCCUUUGACCACCAUGAAACCCCAACCCAAU ..............((((..(((((((..((((((((......))(((((......)))))...)))))).)))))))))))........ ( -29.40) >DroSec_CAF1 50098 90 - 1 UAUUUGCAAUUAUCGGGGAAUUUCGUGCUUGGUCAGCACGAAAGCGAGCGUGCUCCUGCUCCGUUGACCACCAUGAAACCCCAACCCAAU ..............((((..(((((((..((((((((.(....).(((((......))))).)))))))).)))))))))))........ ( -33.10) >DroSim_CAF1 54085 90 - 1 UAUUUGCAAUUAUCGGGGAAUUUCGUGCUUGGUCAGCACGAAAGCGAGCGUGCUCCUGCUCCGUUGACCACCAUGAAACCCCAACCCGAU ...........((((((...(((((((..((((((((.(....).(((((......))))).)))))))).)))))))......)))))) ( -33.20) >DroEre_CAF1 51258 90 - 1 UAUUUGCAAUUAUCGGGGAAUUUCGUGCUUGGUCAGCACAAAAGCAAGCGUGCUCCCGCUCCGUUGACCACCAUGAAACCCCAACCCAAU ..............((((..(((((((..((((((((.(....)..((((......))))..)))))))).)))))))))))........ ( -27.50) >DroYak_CAF1 52455 90 - 1 UAUUUGCAAUUAUCGGGGAAUUUCGUGCUUGGUCAGCACGAAAGCGAGCGUGCUCCUGCUCCGUUGACCACCGUGAAACCCCAACCCAAU ..............((((..(((((((..((((((((.(....).(((((......))))).)))))))).)))))))))))........ ( -29.70) >consensus UAUUUGCAAUUAUCGGGGAAUUUCGUGCUUGGUCAGCACGAAAGCGAGCGUGCUCCUGCUCCGUUGACCACCAUGAAACCCCAACCCAAU ..............((((..(((((((..((((((((.(....).(((((......))))).)))))))).)))))))))))........ (-30.14 = -30.22 + 0.08)

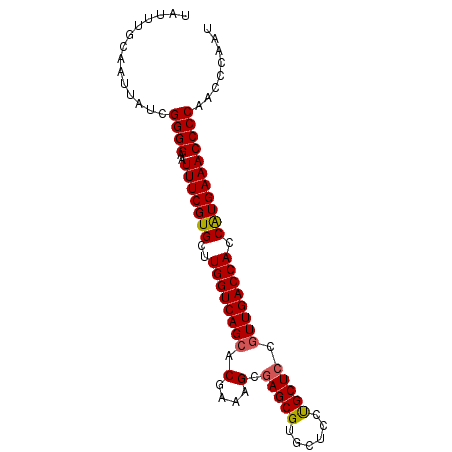
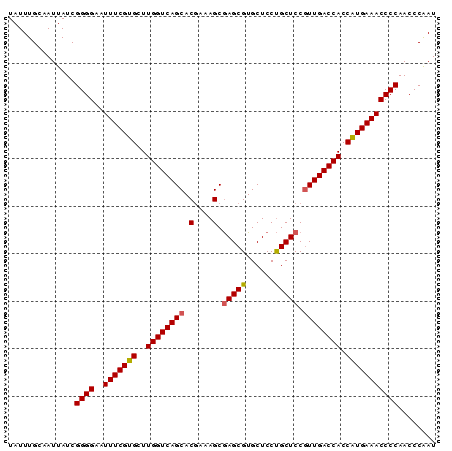
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:48:48 2006