
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 20,233,985 – 20,234,081 |
| Length | 96 |
| Max. P | 0.995368 |

| Location | 20,233,985 – 20,234,081 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 73.68 |
| Mean single sequence MFE | -29.33 |
| Consensus MFE | -21.86 |
| Energy contribution | -21.20 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.75 |
| SVM decision value | 2.57 |
| SVM RNA-class probability | 0.995368 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
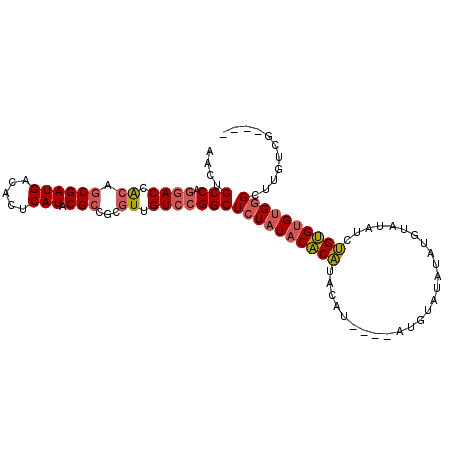
>3R_DroMel_CAF1 20233985 96 + 27905053 ----CGACAAGCCCACACACAGAUAUACAUAUAUACAU----AUGUAUGUGUAUAGAGCCGGACAACGCGGCGUAUGAGUGUCAUCGCUGUAGUCCUGGCAGUU ----.(((..............((((((((((((....----))))))))))))...(((((((...((((((.(((.....))))))))).)))).))).))) ( -32.90) >DroSim_CAF1 49033 100 + 1 ----CGACAAGCCCACACACAGAUAUAAGUAUAUAUAUGUAUGUGUAUGUGUAUAGAGCCGGACAACGCGGCGUAUGAGUGUCAUCGCUGUGGUCCUGGCAGUU ----.(((.....((((.(((.(((((.((....)).))))).))).))))......(((((((..(((((((.(((.....)))))))))))))).))).))) ( -34.10) >DroAna_CAF1 47705 90 + 1 AGAGAGACAAACACACACUCUG-----CACAUAAAC------ACGAUAGAGGAGAGAGCCGGACAAGGCGGCGUAUGAGUGUCAUCGUUGCGGU---GGCAGUU ...((((((..((.((.((((.-----(........------........).)))).((((.......)))))).))..)))).)).((((...---.)))).. ( -20.99) >consensus ____CGACAAGCCCACACACAGAUAUACAUAUAUACAU____AUGUAUGUGUAUAGAGCCGGACAACGCGGCGUAUGAGUGUCAUCGCUGUGGUCCUGGCAGUU .....(((.........((((..........................))))......(((((((...((((((.(((.....))))))))).)))).))).))) (-21.86 = -21.20 + -0.66)


| Location | 20,233,985 – 20,234,081 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 73.68 |
| Mean single sequence MFE | -28.13 |
| Consensus MFE | -16.01 |
| Energy contribution | -17.24 |
| Covariance contribution | 1.23 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.57 |
| SVM decision value | 1.82 |
| SVM RNA-class probability | 0.978679 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20233985 96 - 27905053 AACUGCCAGGACUACAGCGAUGACACUCAUACGCCGCGUUGUCCGGCUCUAUACACAUACAU----AUGUAUAUAUGUAUAUCUGUGUGUGGGCUUGUCG---- ....(((.((((.((.((((((.....))).)))...)).)))))))(((((((((((((((----((....)))))))....)))))))))).......---- ( -31.70) >DroSim_CAF1 49033 100 - 1 AACUGCCAGGACCACAGCGAUGACACUCAUACGCCGCGUUGUCCGGCUCUAUACACAUACACAUACAUAUAUAUACUUAUAUCUGUGUGUGGGCUUGUCG---- ....(((.((((.((.((((((.....))).)))...)).))))))).......(((..((((((((.(((((....))))).))))))))....)))..---- ( -30.90) >DroAna_CAF1 47705 90 - 1 AACUGCC---ACCGCAACGAUGACACUCAUACGCCGCCUUGUCCGGCUCUCUCCUCUAUCGU------GUUUAUGUG-----CAGAGUGUGUGUUUGUCUCUCU ...(((.---...)))..((.((((..(((((((.(((......)))...........((.(------((......)-----)))))))))))..))))))... ( -21.80) >consensus AACUGCCAGGACCACAGCGAUGACACUCAUACGCCGCGUUGUCCGGCUCUAUACACAUACAU____AUGUAUAUAUGUAUAUCUGUGUGUGGGCUUGUCG____ ....(((.((((.((.((((((.....))).)))...)).)))))))((((((((((..........................))))))))))........... (-16.01 = -17.24 + 1.23)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:48:41 2006