| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 2,652,165 – 2,652,263 |
| Length | 98 |
| Max. P | 0.979916 |

| Location | 2,652,165 – 2,652,263 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.67 |
| Mean single sequence MFE | -29.25 |
| Consensus MFE | -21.00 |
| Energy contribution | -20.75 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.783440 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2652165 98 + 27905053 GGCGUGCUGUGGCAAAGUAGACUUUUAAAAGCCAUUUACCGUGGCGUGUUAUUACGGCCAA------CUCAAUUGGCCCGCC----CGCUUUGUU-UUGGAUUUCCAAU----------- ((((.((.(((((((((....)))......(((((.....))))).))))))...((((((------.....)))))).)).----)))).....-((((....)))).----------- ( -28.00) >DroVir_CAF1 8368 89 + 1 GGCGGAAUGUGGCAAAGUAGACUUUUAAAAGCCAUUUACCGUGGCGUGUUAUUAUGGCCAA------GUCAAUUGGCUCGCC----CGCUUU-----UCAACAA---------------- .((((...(((((((((....)))).....)))))...))))((((.........((((((------.....))))))))))----......-----.......---------------- ( -24.60) >DroGri_CAF1 1309 94 + 1 GGCGGUUUGUGGUAAAGUAGACUUUUAAAAGCCAUUUACCGUGGCGUGUUAUU-UGGCCAA------GUCAAUUGGCCCGCC----CGGUAU-----UUAACU-CACUA---------GA ...(((((.....((((....))))...)))))...(((((.((((.......-.((((((------.....))))))))))----))))).-----......-.....---------.. ( -28.50) >DroMoj_CAF1 16077 96 + 1 GGCGGACUGUGGCAAAGUAGACUUUUAAAAGCCAUUUACCGUGGCGUGUUAUUAUGGCCAA------CUCAAUUGGCCCGCC----CGGUAU-----UCAACAACAAUA---------AA ...(((..(((((((((....)))).....))))).(((((.((((.........((((((------.....))))))))))----))))))-----))..........---------.. ( -27.00) >DroAna_CAF1 899 110 + 1 GGCGUGCUACGGCAAAGUAGACUUUUAAAAGCCAUUUACCGUGGCGUGUUAUUACGGCCAA------CUCAAUUGGCCCGCC----CGGUUUGUUUUUGGCUUUCCAAUCCAAUACCAGG ((..(((....))).............(((((((...((((.((((((....)))((((((------.....)))))).)))----)))).......)))))))...........))... ( -34.90) >DroPer_CAF1 25776 110 + 1 GGCGUGCUGUGGCAAAGUAGACUUUUAAAAGCCAUUUACCGUGGCGUGUUAUUAUGGCCAAGUCCAAGUCAAUUGGCCCGCCCGCCCGCCUCGUA-UUCGCUGCCAAAG---------AG ((((.((.(((((((((....)))).....)))))....((.((((.((......((((((...........)))))).....)).)))).))..-...))))))....---------.. ( -32.50) >consensus GGCGGGCUGUGGCAAAGUAGACUUUUAAAAGCCAUUUACCGUGGCGUGUUAUUAUGGCCAA______CUCAAUUGGCCCGCC____CGCUUU_____UCAACUACAAAA___________ ((((....(((((((((....)))......(((((.....))))).))))))...((((((...........))))))))))...................................... (-21.00 = -20.75 + -0.25)

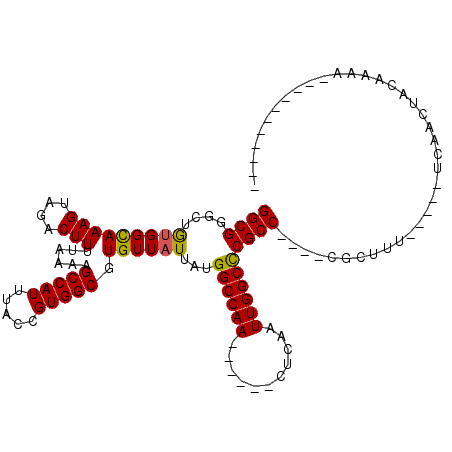
| Location | 2,652,165 – 2,652,263 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.67 |
| Mean single sequence MFE | -31.07 |
| Consensus MFE | -22.97 |
| Energy contribution | -23.00 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.85 |
| SVM RNA-class probability | 0.979916 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
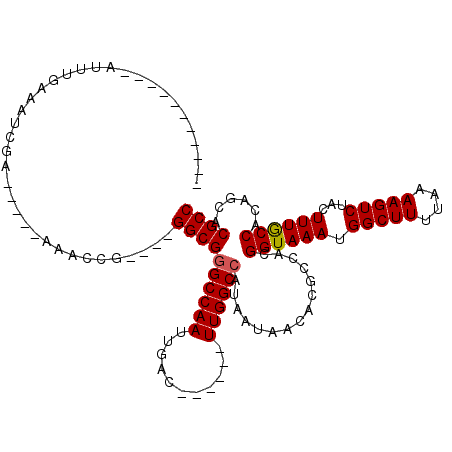
>3R_DroMel_CAF1 2652165 98 - 27905053 -----------AUUGGAAAUCCAA-AACAAAGCG----GGCGGGCCAAUUGAG------UUGGCCGUAAUAACACGCCACGGUAAAUGGCUUUUAAAAGUCUACUUUGCCACAGCACGCC -----------.((((....))))-......(((----(((((((((((...)------))))))((....)).))))..((((((.(((((....)))))...))))))......))). ( -34.00) >DroVir_CAF1 8368 89 - 1 ----------------UUGUUGA-----AAAGCG----GGCGAGCCAAUUGAC------UUGGCCAUAAUAACACGCCACGGUAAAUGGCUUUUAAAAGUCUACUUUGCCACAUUCCGCC ----------------.((((..-----..))))----((((.(((((.....------)))))................((((((.(((((....)))))...))))))......)))) ( -23.20) >DroGri_CAF1 1309 94 - 1 UC---------UAGUG-AGUUAA-----AUACCG----GGCGGGCCAAUUGAC------UUGGCCA-AAUAACACGCCACGGUAAAUGGCUUUUAAAAGUCUACUUUACCACAAACCGCC ..---------..(((-(((((.-----.(((((----((((((((((.....------)))))).-.......)))).)))))..)))))....((((....))))..)))........ ( -27.80) >DroMoj_CAF1 16077 96 - 1 UU---------UAUUGUUGUUGA-----AUACCG----GGCGGGCCAAUUGAG------UUGGCCAUAAUAACACGCCACGGUAAAUGGCUUUUAAAAGUCUACUUUGCCACAGUCCGCC ..---------.(((((.((.((-----.(((((----(((((((((((...)------)))))).........)))).)))))...(((((....)))))...)).)).)))))..... ( -27.60) >DroAna_CAF1 899 110 - 1 CCUGGUAUUGGAUUGGAAAGCCAAAAACAAACCG----GGCGGGCCAAUUGAG------UUGGCCGUAAUAACACGCCACGGUAAAUGGCUUUUAAAAGUCUACUUUGCCGUAGCACGCC ...((((.((((((.((((((((.......((((----(((((((((((...)------))))))((....)).)))).))))...))))))))...))))))...)))).......... ( -41.50) >DroPer_CAF1 25776 110 - 1 CU---------CUUUGGCAGCGAA-UACGAGGCGGGCGGGCGGGCCAAUUGACUUGGACUUGGCCAUAAUAACACGCCACGGUAAAUGGCUUUUAAAAGUCUACUUUGCCACAGCACGCC ..---------.....((..((..-..))..)).((((((((((((((...........)))))).........))))..((((((.(((((....)))))...))))))......)))) ( -32.30) >consensus ___________AUUUGAAAUCGA_____AAACCG____GGCGGGCCAAUUGAC______UUGGCCAUAAUAACACGCCACGGUAAAUGGCUUUUAAAAGUCUACUUUGCCACAGCACGCC ......................................((((((((((...........))))))...............((((((.(((((....)))))...))))))......)))) (-22.97 = -23.00 + 0.03)

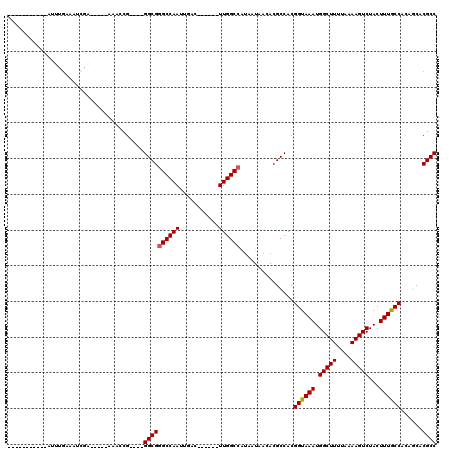
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:51:19 2006