
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 20,221,880 – 20,222,024 |
| Length | 144 |
| Max. P | 0.792687 |

| Location | 20,221,880 – 20,221,996 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.52 |
| Mean single sequence MFE | -28.43 |
| Consensus MFE | -24.04 |
| Energy contribution | -24.04 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.765149 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
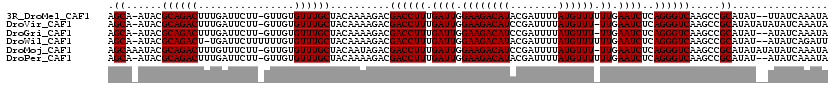
>3R_DroMel_CAF1 20221880 116 + 27905053 AGCA-AUACGCAGACUUUGAUUCUU-GUUGUGUUUGCUACAAAAGACGACCUUUGAUUGGAAGACAUACGAUUUUAUGUUUUUUGAAUCUCAGGGUCAAGCCGCAUAU--UUAUCAAAUA .((.-....((((((..((((....-)))).))))))..........((((((.(((..(((((((((......)))))))))..).))..)))))).....))....--.......... ( -27.70) >DroVir_CAF1 42887 117 + 1 AGCA-AUACGCAGACUUUGAUUCUU-GUUGUGUUUGCUACAAAAGACGACCUUUGAUUGGAAGACAUCCGAUUUUAUGUUU-UUGAAUCUCAGGGUCAAGCCGCAUAUAUAUAUCAAAUA .((.-....((((((..((((....-)))).))))))..........((((((.((((.((((((((........))))))-)).))))..)))))).....))................ ( -28.20) >DroGri_CAF1 46579 115 + 1 AGCA-AUACGCAGACUUUGAUUCUU-GUUGUGUUUGCUACAAAAGACGACCUUUGAUUGGAAGACAUCCGAUUUUAUGUUU-UUGAAUCUCAGGGUCAAGCCGCAUAU--AUAUCAAAUA .((.-....((((((..((((....-)))).))))))..........((((((.((((.((((((((........))))))-)).))))..)))))).....))....--.......... ( -28.20) >DroWil_CAF1 34124 116 + 1 AGCA-AUACGCAGACU-UGAUUCUUUUUUGUGUUUGCUACAAAAGACGACCUUUGAUUGGAAGACAUACGAUUUUAUGUUUUUUGAAUCUCAGGGUCAAGCCGCAUAU--AUAUCAGAUU ....-....((.(.((-(((((((((((((((.....))))))))).))(((..(((..(((((((((......)))))))))..).))..)))))))))).))....--.......... ( -28.60) >DroMoj_CAF1 40562 118 + 1 AGCAAAUACGCAGACUUUGUUUCUU-GUUGUGUUUGCUACAAUAGACGACCUUUGAUUGGAAGACAUCCGAUUUUAUGUUU-UUGAAUCUCAGGGUCAAGCCGCAUAUAUAUAUCAAAUA ((((((((((((((.......)).)-).)))))))))).........((((((.((((.((((((((........))))))-)).))))..))))))....................... ( -30.20) >DroPer_CAF1 37388 116 + 1 AGCA-AUACGCAGACUUUGAUUCUU-GUUGUGUUUGCUACAAAAGACGACCUUUGAUUGGAAGACAUACGAUUUUAUGUUUUUUGAAUCUCAGGGUCAAGCCGCAUAU--AUAUCAAAUA .((.-....((((((..((((....-)))).))))))..........((((((.(((..(((((((((......)))))))))..).))..)))))).....))....--.......... ( -27.70) >consensus AGCA_AUACGCAGACUUUGAUUCUU_GUUGUGUUUGCUACAAAAGACGACCUUUGAUUGGAAGACAUACGAUUUUAUGUUU_UUGAAUCUCAGGGUCAAGCCGCAUAU__AUAUCAAAUA .((......((((((................))))))..........((((((.((((.((((((((........)))))).)).))))..)))))).....))................ (-24.04 = -24.04 + -0.00)

| Location | 20,221,918 – 20,222,024 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 78.09 |
| Mean single sequence MFE | -19.60 |
| Consensus MFE | -16.40 |
| Energy contribution | -17.07 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.18 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.792687 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
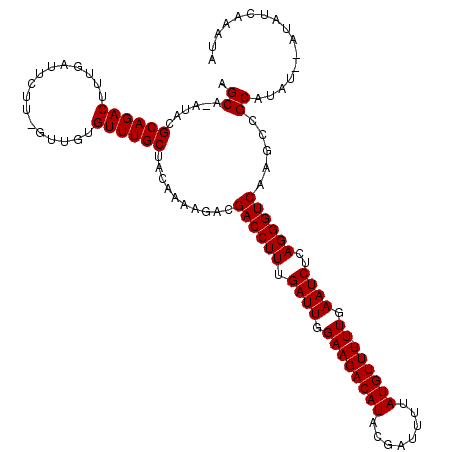
>3R_DroMel_CAF1 20221918 106 + 27905053 AAAAGACGACCUUUGAUUGGAAGACAUACGAUUUUAUGUUUUUUGAAUCUCAGGGUCAAGCCGCAUAU--UUAUCAAAUACCUCA-AU--------UUCCUGUUUACAUCGUAAUUA .......((((((.(((..(((((((((......)))))))))..).))..))))))...........--...............-..--------..................... ( -18.30) >DroVir_CAF1 42925 85 + 1 AAAAGACGACCUUUGAUUGGAAGACAUCCGAUUUUAUGUUU-UUGAAUCUCAGGGUCAAGCCGCAUAUAUAUAUCAAAUACUUAUU------------------------------- .......((((((.((((.((((((((........))))))-)).))))..)))))).............................------------------------------- ( -18.80) >DroPse_CAF1 38850 106 + 1 AAAAGACGACCUUUGAUUGGAAGACAUACGAUUUUAUGUUUUUUGAAUCUCAGGGUCAAGCCGCAUAU--AUAUCAAAUACGUCA-AU--------UUCUUGAUUACACCAUAAGUU ....(((((((((.(((..(((((((((......)))))))))..).))..))))))...........--...........((((-(.--------...)))))..........))) ( -20.30) >DroGri_CAF1 46617 83 + 1 AAAAGACGACCUUUGAUUGGAAGACAUCCGAUUUUAUGUUU-UUGAAUCUCAGGGUCAAGCCGCAUAU--AUAUCAAAUACUUAGU------------------------------- .......((((((.((((.((((((((........))))))-)).))))..))))))...........--................------------------------------- ( -18.80) >DroWil_CAF1 34162 114 + 1 AAAAGACGACCUUUGAUUGGAAGACAUACGAUUUUAUGUUUUUUGAAUCUCAGGGUCAAGCCGCAUAU--AUAUCAGAUUACUUA-GUUUUUCAGUUUUGUUUUUGUUUAAUAACUA .((((((((.....(((((((((((....(((..(((((..(((((.((....)))))))..))))).--..)))((....))..-)))))))))))))))))))............ ( -21.10) >DroPer_CAF1 37426 106 + 1 AAAAGACGACCUUUGAUUGGAAGACAUACGAUUUUAUGUUUUUUGAAUCUCAGGGUCAAGCCGCAUAU--AUAUCAAAUACGUCA-AU--------UUCUUGAUUACACCAUAAGUU ....(((((((((.(((..(((((((((......)))))))))..).))..))))))...........--...........((((-(.--------...)))))..........))) ( -20.30) >consensus AAAAGACGACCUUUGAUUGGAAGACAUACGAUUUUAUGUUUUUUGAAUCUCAGGGUCAAGCCGCAUAU__AUAUCAAAUACCUCA_AU________UUC_UG_UUACA_CAUAA_U_ .......((((((.(((..(((((((((......)))))))))..).))..))))))............................................................ (-16.40 = -17.07 + 0.67)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:48:32 2006