| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 20,214,572 – 20,214,679 |
| Length | 107 |
| Max. P | 0.744766 |

| Location | 20,214,572 – 20,214,679 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.01 |
| Mean single sequence MFE | -25.70 |
| Consensus MFE | -21.63 |
| Energy contribution | -21.35 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.744766 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
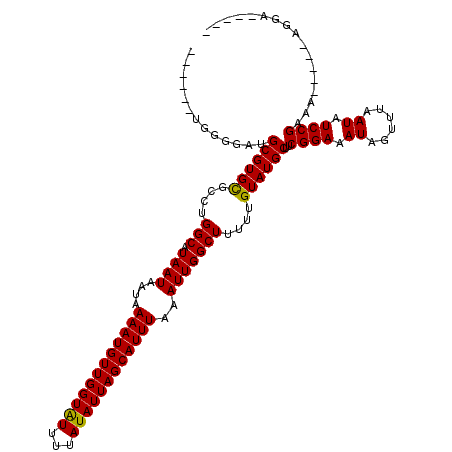
>3R_DroMel_CAF1 20214572 107 - 27905053 --------GCGAUGCGUGCUCAUGGCAUAAUAAUAAAAUGUUGGUAUUUUAUAUUAGCAUUUAAAUUGGCUUUUUGUAUGCUUCGGAAAUAGUUUAAUAUCCGUAG-----AGGACCGUG --------.(((.((((((....(((.((((....((((((((((((...))))))))))))..)))))))....)))))).)))..............(((....-----.)))..... ( -25.50) >DroVir_CAF1 34920 103 - 1 ------UGUGAAUGCGUGUGGCUGGCAUAAUAAUAAAAUGUUGGUGUUUUAUAUUAGCAUUUAAAUUGGCUUUUUGUAUGCUUCGGAAAUAGUUUAAUAUCCGAAA-----AGU------ ------.......(((((..(..(((.((((....((((((((((((...))))))))))))..)))))))..)..)))))((((((.((......)).)))))).-----...------ ( -25.20) >DroWil_CAF1 26496 110 - 1 AAUGUAUGUGGCAGCGUGCGCCUGGCAUAAUAAUAAAAUGUUGGUAUUUUAUAUUAGCAUUUAAAUUGGCUUUUUGUAUGCUUCGGAAAUAGUUUAAUAUCCGAAA-----AAGA----- ..(((.....)))(((((((...(((.((((....((((((((((((...))))))))))))..)))))))...)))))))((((((.((......)).)))))).-----....----- ( -25.90) >DroMoj_CAF1 30556 103 - 1 ------UGUGAAUGCGUGCGGCUGGCAUAAUAAUAAAAUGUUGGUAUUUUAUAUUAGCAUUUAAAUUGGCUUUUUGUAUGCUUCGGAAAUAGUUUAAUAUCCGAAA-----AGU------ ------.......((((((((..(((.((((....((((((((((((...))))))))))))..)))))))..))))))))((((((.((......)).)))))).-----...------ ( -27.10) >DroAna_CAF1 26470 108 - 1 ---GU-AGGGGAUGCGUGCUCUUGGCAUAAUAAUAAAAUGUUGGUAUUUUAUAUUAGCAUUUAAAUUGGCUUUUUGUAUGCUUCGGAAAUAGUUUAAUAUCCGCAG-----AGGAAU--- ---..-....((.((((((....(((.((((....((((((((((((...))))))))))))..)))))))....)))))).))...............(((....-----.)))..--- ( -23.60) >DroPer_CAF1 29779 112 - 1 ------AGGGGGUGCGUGCUCUUGGCAUAAUAAUAAAAUGUUGGUAUUUUAUAUUAGCAUUUAAAUUGGCUUUUUGUAUGCUUCGGAAAUAGUUUAAUAUCCGUAGGGGCGAGGACUA-- ------....(((.(.(((((((((((((.((((.((((((((((((...))))))))))))..))))........)))))..((((.((......)).)))))))))))).).))).-- ( -26.90) >consensus ______UGGGGAUGCGUGCGCCUGGCAUAAUAAUAAAAUGUUGGUAUUUUAUAUUAGCAUUUAAAUUGGCUUUUUGUAUGCUUCGGAAAUAGUUUAAUAUCCGAAA_____AGGA_____ .............((((((....(((.((((....((((((((((((...))))))))))))..)))))))....))))))..((((.((......)).))))................. (-21.63 = -21.35 + -0.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:48:29 2006