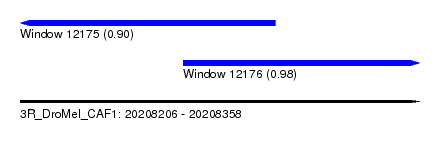
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 20,208,206 – 20,208,358 |
| Length | 152 |
| Max. P | 0.984117 |

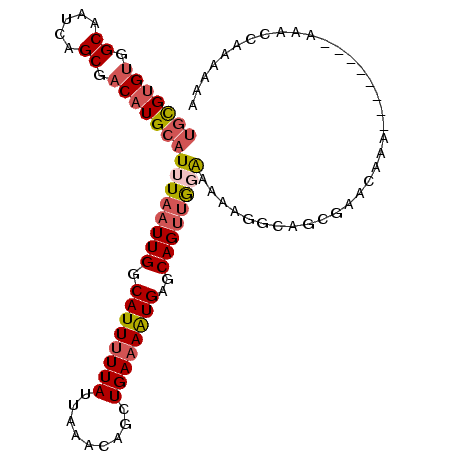
| Location | 20,208,206 – 20,208,303 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 81.79 |
| Mean single sequence MFE | -23.35 |
| Consensus MFE | -17.33 |
| Energy contribution | -18.17 |
| Covariance contribution | 0.83 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.903939 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20208206 97 - 27905053 UGCGUGUGGCAAUCAGCGACAUGCAUUUAAUUGGCAUUUUUAUUAAACAGCUGAAAAUGAGCAGUUGGGAAAAGGCAGCGAACAAA--------AAACCAAAAAA (((((((.((.....)).)))))))........((.(((((......((((((........)))))).))))).))..........--------........... ( -22.90) >DroSec_CAF1 19473 97 - 1 UGCGUGUGGCAAUCAGCGACAUGCAUUUAAUUGGCAUUUUUAUUAAACAGCUGAAAAUGAGCAGUUGCAAAGAGGCAGCGAACAAA--------AAACCAAAAAA (((((((.((.....)).))))))).....((((((((((((.........))))))))....(((((......))))).......--------...)))).... ( -26.10) >DroSim_CAF1 19485 97 - 1 UGCGUGUGGCAAUCAGCGACAUGCAUUUAAUUGGCAUUUUUAUUAAACAGCUGAAAAUGAGCAGUUGCAAAGAGGCAGCGAACAAA--------AAACCAAAAAA (((((((.((.....)).))))))).....((((((((((((.........))))))))....(((((......))))).......--------...)))).... ( -26.10) >DroEre_CAF1 19709 88 - 1 UGCGUGUGGCAAUCAGCGACAUGCAUUUAAUUGGCAUUUUUAUUAAACAGCUGAAAAUGAGCAGUUGGGCAAAGGCUGCA-----------------CUAAAAAA (((((((.((.....)).))))))).....((((((((((((.........)))))))).((((((.......)))))).-----------------)))).... ( -24.70) >DroYak_CAF1 20155 105 - 1 UGCGUGUGGCAAUCAGCGACAUGCAUUUAAUUGGCAUUUUUAUUAAACAGCUGAAAAUGAGCAGUUGGGCAAAAGCUGCAAACAAAUACAAUCAACACGAAAAAA (((((((.((.....)).)))))))....((((.((((((((.........)))))))).((((((.......)))))).........))))............. ( -25.70) >DroPer_CAF1 23134 88 - 1 -UUGCGGUGCAAUCAGCAACAUGCAUUUAAUUGGCAUUAUUAUUAAACAGCUGAAAGUGAGCAGCAAGAAAAAAGG--CGGAUAAG--------GA--C----AA -.((((.(((.....)))...))))....(((.((.(((....)))...((((........))))..........)--).)))...--------..--.----.. ( -14.60) >consensus UGCGUGUGGCAAUCAGCGACAUGCAUUUAAUUGGCAUUUUUAUUAAACAGCUGAAAAUGAGCAGUUGGAAAAAGGCAGCGAACAAA________AAACCAAAAAA (((((((.((.....)).)))))))((((((((.((((((((.........))))))))..)))))))).................................... (-17.33 = -18.17 + 0.83)

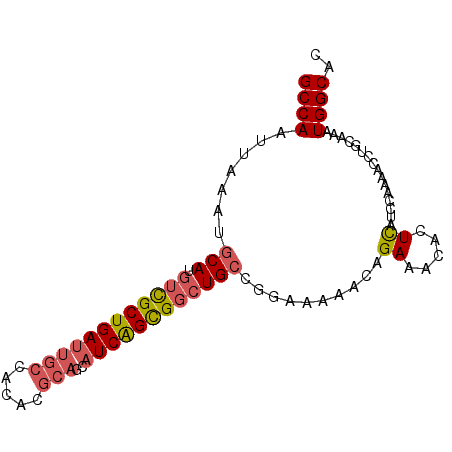
| Location | 20,208,268 – 20,208,358 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 86.33 |
| Mean single sequence MFE | -25.67 |
| Consensus MFE | -19.04 |
| Energy contribution | -19.80 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.92 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.96 |
| SVM RNA-class probability | 0.984117 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20208268 90 + 27905053 GCCAAUUAAAUGCAUGUCGCUGAUUGCCACACGCAGCAUCAGUGGCUGCCGGAAAAACAGAAACACUCAUCCAAAACCUGCAAAUGGCAC ((((.......(((.(((((((((.((........)))))))))))))).(((................)))............)))).. ( -24.79) >DroSec_CAF1 19535 90 + 1 GCCAAUUAAAUGCAUGUCGCUGAUUGCCACACGCAGCAUCAGCGGCUGCCGGACAAACAGAAACACUCAUCCAAAACCUGCAAAUGGCAC ((((.......(((.(((((((((.((........)))))))))))))).(((................)))............)))).. ( -26.49) >DroSim_CAF1 19547 90 + 1 GCCAAUUAAAUGCAUGUCGCUGAUUGCCACACGCAGCAUCAGCGGCUGCCGGAAAAACAGAAACACUCAUCCAAAACCUGCAAAUGGCAC ((((.......(((.(((((((((.((........)))))))))))))).(((................)))............)))).. ( -26.69) >DroEre_CAF1 19762 90 + 1 GCCAAUUAAAUGCAUGUCGCUGAUUGCCACACGCAGGAUCAGCGGCUGCCGGAAAAACAGAAACACUCAUCCAAAACCUGCAAAUGGCAC ((((.......(((.((((((((((((.....)))..)))))))))))).(((................)))............)))).. ( -26.59) >DroAna_CAF1 20758 82 + 1 GCCAAUUAAAUGCACGGUGCUGAUUGCCAU--------UCGGCGGGUGGAAAAAAUAACGAAGCACUUUGCAAAGUCCUGCAAAUGGCAC ((((......((((.((((((.(((.((((--------(.....)))))....))).....)))))).))))..(.....)...)))).. ( -23.80) >consensus GCCAAUUAAAUGCAUGUCGCUGAUUGCCACACGCAGCAUCAGCGGCUGCCGGAAAAACAGAAACACUCAUCCAAAACCUGCAAAUGGCAC ((((.......(((.((((((((((((.....)))..))))))))))))..........((.....))................)))).. (-19.04 = -19.80 + 0.76)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:48:28 2006