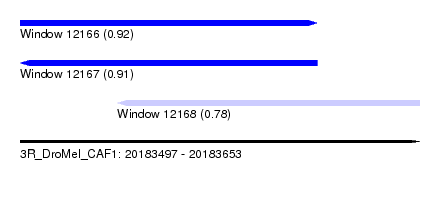
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 20,183,497 – 20,183,653 |
| Length | 156 |
| Max. P | 0.917887 |

| Location | 20,183,497 – 20,183,613 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.77 |
| Mean single sequence MFE | -35.53 |
| Consensus MFE | -28.50 |
| Energy contribution | -27.33 |
| Covariance contribution | -1.16 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.917887 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
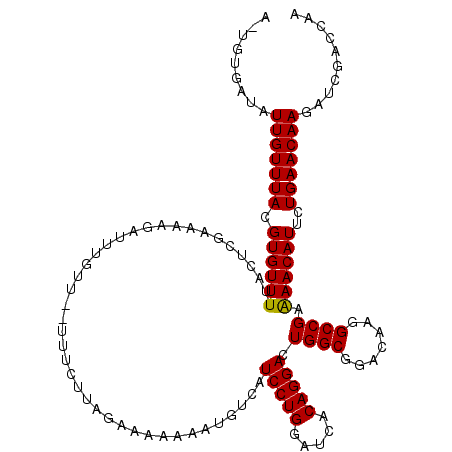
>3R_DroMel_CAF1 20183497 116 + 27905053 C--CUGUGGCGUUCUCUGUCUGCAGGGCCGUUGCCGCAUUUUGGUCCAGCUUGUUCAAAAUGUUUUCGGCCUUGUCCGCCAGUCCUGUGAUCCAGGAUGACAUAUUUUUCUAAGAAA--A .--...(((((..........(((((((((.....(((((((((..........)))))))))...))))))))).)))))((((((.....))))))...................--. ( -35.60) >DroPse_CAF1 9224 109 + 1 CAACGCUG---UUUCCAG-UUGCAGGGCCGUGGCCGCAUUCUGGUCGAUCUUGUUCAGAAUGUUCUCGGCCUUAUCUGCCAAUCCUGUGAUCCAGGAUGACAUUU-----UAAACGA--A ....((((---....)))-).(((((((((.((..((((((((..((....))..)))))))))).)))))....))))..((((((.....)))))).......-----.......--. ( -33.90) >DroEre_CAF1 8471 102 + 1 C--CGGUGGAGUUCUCUGUCUGCAGGGCCGUUGCCGCAUUUUGGUCCAGCUUGUUCAAAAUGUUUUCGGCCUUAUCUGCCAGUCCUGUGAUCCAGGAUGACAUA---------------- .--.((..((.....(((....)))(((((.....(((((((((..........)))))))))...)))))...))..)).((((((.....))))))......---------------- ( -34.40) >DroYak_CAF1 8435 116 + 1 C--CGGUGGAGUUCUCUGUUUGGAGGGCUGUUGCCGCAUUUUGGUCCAGCUUGUUCAAGAUGUUUUCGGCCUUGUCCGCCAGUCCUGUGAUCCAGGAUGACAUAUUUUUCUAAAACA--A .--.(((((((((((((....)))))))....((((((((((((..........))))))))....))))....)))))).((((((.....))))))...................--. ( -36.10) >DroAna_CAF1 8199 118 + 1 C--CGGCUUGAUUAUCCGUCUGUAGGGCUGUCGCCGCAUUCUGAUCGAUCUUGUUCAGAAUGUUUUCGGCCUUGUCCGCCAAUCCUGUUAUCCAGGAUGACAUUUUUUGCUCGGAAAUAA (--((((..((......(((((..((((.((((..(((((((((.((....)).)))))))))...))))...))))..))((((((.....)))))))))...))..)).)))...... ( -36.60) >DroPer_CAF1 9243 112 + 1 CAACGCUGUCGUUUCCAG-UUGCAGGGCCGUGGCCGCAUUCUGGUCGAUCUUGUUCAGAAUGUUCUCGGCCUUAUCUGCCAAUCCUGUGAUCCAGGAUGACAUUU-----UAAACGA--A .......((((...((((-.(((..(((....))))))..)))).)))).(((((.((((((((...(((.......))).((((((.....)))))))))))))-----).)))))--. ( -36.60) >consensus C__CGGUGGAGUUCUCUGUCUGCAGGGCCGUUGCCGCAUUCUGGUCCAGCUUGUUCAAAAUGUUUUCGGCCUUAUCCGCCAAUCCUGUGAUCCAGGAUGACAUAU_____UAAAAAA__A .................(((...(((((((.....(((((((((..........)))))))))...)))))))........((((((.....)))))))))................... (-28.50 = -27.33 + -1.16)



| Location | 20,183,497 – 20,183,613 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.77 |
| Mean single sequence MFE | -32.63 |
| Consensus MFE | -24.65 |
| Energy contribution | -24.40 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.908069 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20183497 116 - 27905053 U--UUUCUUAGAAAAAUAUGUCAUCCUGGAUCACAGGACUGGCGGACAAGGCCGAAAACAUUUUGAACAAGCUGGACCAAAAUGCGGCAACGGCCCUGCAGACAGAGAACGCCACAG--G .--....................(((((.....))))).(((((..(..((((.....(((((((..(.....)...))))))).(....)))))(((....))).)..)))))...--. ( -30.80) >DroPse_CAF1 9224 109 - 1 U--UCGUUUA-----AAAUGUCAUCCUGGAUCACAGGAUUGGCAGAUAAGGCCGAGAACAUUCUGAACAAGAUCGACCAGAAUGCGGCCACGGCCCUGCAA-CUGGAAA---CAGCGUUG .--((((...-----...((((((((((.....))))).))))).....(((((....(((((((..(......)..))))))))))))))))........-(((....---)))..... ( -35.60) >DroEre_CAF1 8471 102 - 1 ----------------UAUGUCAUCCUGGAUCACAGGACUGGCAGAUAAGGCCGAAAACAUUUUGAACAAGCUGGACCAAAAUGCGGCAACGGCCCUGCAGACAGAGAACUCCACCG--G ----------------..((((.(((((.....)))))...((((....((((.....(((((((..(.....)...))))))).(....))))))))).)))).............--. ( -29.90) >DroYak_CAF1 8435 116 - 1 U--UGUUUUAGAAAAAUAUGUCAUCCUGGAUCACAGGACUGGCGGACAAGGCCGAAAACAUCUUGAACAAGCUGGACCAAAAUGCGGCAACAGCCCUCCAAACAGAGAACUCCACCG--G (--(((((.(((.......(((((((((.....))))).))))((......)).......))).))))))(.((((.........(((....)))(((......)))...)))).).--. ( -28.80) >DroAna_CAF1 8199 118 - 1 UUAUUUCCGAGCAAAAAAUGUCAUCCUGGAUAACAGGAUUGGCGGACAAGGCCGAAAACAUUCUGAACAAGAUCGAUCAGAAUGCGGCGACAGCCCUACAGACGGAUAAUCAAGCCG--G ......(((.((......((((((((((.....))))).)))))((.....(((....((((((((.(......).)))))))).(((....))).......)))....))..))))--) ( -35.80) >DroPer_CAF1 9243 112 - 1 U--UCGUUUA-----AAAUGUCAUCCUGGAUCACAGGAUUGGCAGAUAAGGCCGAGAACAUUCUGAACAAGAUCGACCAGAAUGCGGCCACGGCCCUGCAA-CUGGAAACGACAGCGUUG .--.......-----...((((((((((.....))))).))))).....(((((....(((((((..(......)..)))))))))))).((((.(((...-.((....)).))).)))) ( -34.90) >consensus U__UCUUUUA_____AAAUGUCAUCCUGGAUCACAGGACUGGCAGACAAGGCCGAAAACAUUCUGAACAAGAUCGACCAAAAUGCGGCAACGGCCCUGCAAACAGAGAACGCCACCG__G ..................((((((((((.....))))).))))).....(((((....(((((((............)))))))......)))))......................... (-24.65 = -24.40 + -0.25)



| Location | 20,183,535 – 20,183,653 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.14 |
| Mean single sequence MFE | -28.82 |
| Consensus MFE | -19.91 |
| Energy contribution | -19.68 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.777400 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20183535 118 - 27905053 AGUGUGAUAUUGUUUACGUGUUUUACUCGAAGAGAUUUGUU--UUUCUUAGAAAAAUAUGUCAUCCUGGAUCACAGGACUGGCGGACAAGGCCGAAAACAUUUUGAACAAGCUGGACCAA .((.((...(((((((.(((((((.........(((.((((--((((...)))))))).))).(((((.....))))).((((.......)))))))))))..)))))))..)).))... ( -27.30) >DroPse_CAF1 9260 102 - 1 ------AUUUUGUUUACGUGUUUCAC-----AAGAUUUAUU--UCGUUUA-----AAAUGUCAUCCUGGAUCACAGGAUUGGCAGAUAAGGCCGAGAACAUUCUGAACAAGAUCGACCAG ------((((((((((.(((((((..-----..((......--))((((.-----...((((((((((.....))))).)))))....)))).))).))))..))))))))))....... ( -25.50) >DroSim_CAF1 8899 118 - 1 AGUGUUAUAUUGUUUACGUGUUUUGCUCGAAGAGAUUUGUU--UUUCUUAGAAAAAUAUGUCAUCCUGGAUCACAGGACUGGCGGACAAGGCCGAAAACAUUUUGAACAAGCUGGACCAA ..((((....(((((.((.((((((..((.((.(((.((((--((((...)))))))).))).(((((.....)))))))..))..)))))))).))))).....))))........... ( -32.20) >DroYak_CAF1 8473 118 - 1 ACUGUGAUAUUGUUUACGUGUUUUGCUCGCAGAGAUUAGUU--UGUUUUAGAAAAAUAUGUCAUCCUGGAUCACAGGACUGGCGGACAAGGCCGAAAACAUCUUGAACAAGCUGGACCAA .((((((...................))))))...((((((--(((((.(((.......(((((((((.....))))).))))((......)).......))).)))))))))))..... ( -31.71) >DroAna_CAF1 8237 119 - 1 UAAUAGAUAUUGUUUACGUGUUUGG-UUGACAAGAUUGUUUUAUUUCCGAGCAAAAAAUGUCAUCCUGGAUAACAGGAUUGGCGGACAAGGCCGAAAACAUUCUGAACAAGAUCGAUCAG .....(((.(((((((.((((((..-.(((((...((((((.......))))))....)))))(((((.....)))))(((((.......)))))))))))..))))))).)))...... ( -31.10) >DroPer_CAF1 9282 102 - 1 ------UUUUUGUUUACGUGUUUCAC-----AAGAUUUAUU--UCGUUUA-----AAAUGUCAUCCUGGAUCACAGGAUUGGCAGAUAAGGCCGAGAACAUUCUGAACAAGAUCGACCAG ------.(((((((((.(((((((..-----..((......--))((((.-----...((((((((((.....))))).)))))....)))).))).))))..)))))))))........ ( -25.10) >consensus A_UGUGAUAUUGUUUACGUGUUUUACUCGAAAAGAUUUGUU__UUUCUUAGAAAAAAAUGUCAUCCUGGAUCACAGGACUGGCGGACAAGGCCGAAAACAUUCUGAACAAGAUCGACCAA .........(((((((.((((((........................................(((((.....))))).((((.......)))).))))))..))))))).......... (-19.91 = -19.68 + -0.22)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:48:20 2006