
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 20,167,189 – 20,167,318 |
| Length | 129 |
| Max. P | 0.671430 |

| Location | 20,167,189 – 20,167,289 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 74.28 |
| Mean single sequence MFE | -33.33 |
| Consensus MFE | -18.48 |
| Energy contribution | -18.48 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.671430 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
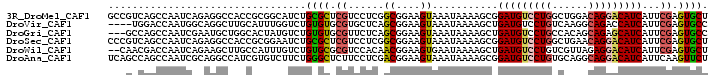
>3R_DroMel_CAF1 20167189 100 + 27905053 GCCGUCAGCCAAUCAGAGGCCACCGCGGCAUCUGCGCUCGUCCUCGGCGGAAGUAAAUAAAAGCGGAUGUCCUGGCUGGACAGGACAUCAUUCGAGUGCU (((((..(((.......)))....)))))....(((((((......((....))...........(((((((((......)))))))))...))))))). ( -42.50) >DroVir_CAF1 8895 96 + 1 ----UGGACCAAUGGCAGGCUUGCAUUUGGUCUGUGUGCGUGCUCAGCGGAAGUAAAUAAAAGCUGAUGUCCUGUCAAGGCAGACCAUCAUUCGAGUGCC ----.(((((((..((......))..)))))))......(..(((.((....))..........(((((..((((....))))..)))))...)))..). ( -32.60) >DroGri_CAF1 10110 97 + 1 ---GCCAGCCAAUCGAAUGCUGGCACUAUGUCUGUGUGCGUUCUCAGCGGAAGUAAAUAAAAGCUGAUGUCCUGCCACAGCAGAGCAUCAUUCGAGUGCC ---((((((.........))))))...............((.(((.((....))..........(((((..((((....))))..)))))...))).)). ( -30.30) >DroSec_CAF1 7941 100 + 1 CCCGUCAGCCAAUCAGAGGCCACCGCGGAAUCUGCGCUCGUCCUCGGCGGAAGUAAAUAAAAGCGGAUGUCCUGGCUGAACAGGACAUCAUUCGAGUGCU .((((..(((.......)))....)))).....(((((((......((....))...........(((((((((......)))))))))...))))))). ( -38.80) >DroWil_CAF1 8356 98 + 1 --CAACGACCAAUCAGAAGCUUGCCAUUUGUCUGUGCGCGUCCACAACGGAAGUGAAUAAAAGCUGAUGUCCUGUCGUUAGAGGACAUCAUUCGAGUGCU --...(((.........((((((((((......))).)).......((....))......)))))((((((((........))))))))..)))...... ( -25.10) >DroAna_CAF1 7911 100 + 1 UCAGCCAGCCAAUCGCAGGCCAUCGUGUCUUCUGGGCUCUUCCUCGACGGAAGUAAAUAAAAGCGGAUGUCCUGUGCAGGCAGGACAUCAUUCAAGUUCU ...((.((((....(.((((......)))).)..))))(((((.....))))).........)).((((((((((....))))))))))........... ( -30.70) >consensus __CGUCAGCCAAUCAGAGGCCAGCGCGUCGUCUGUGCGCGUCCUCAGCGGAAGUAAAUAAAAGCGGAUGUCCUGUCAAGACAGGACAUCAUUCGAGUGCU .................................((((.((......((....))...........(((((((((......)))))))))...)).)))). (-18.48 = -18.48 + 0.00)

| Location | 20,167,209 – 20,167,318 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 75.37 |
| Mean single sequence MFE | -32.97 |
| Consensus MFE | -20.44 |
| Energy contribution | -21.05 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.612402 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
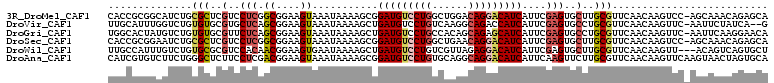
>3R_DroMel_CAF1 20167209 109 + 27905053 CACCGCGGCAUCUGCGCUCGUCCUCGGCGGAAGUAAAUAAAAGCGGAUGUCCUGGCUGGACAGGACAUCAUUCGAGUGCUUGCGUUCAACAAGUCC-AGCAAACAGAGCA ....((((...))))(((((..((((((....))...........(((((((((......)))))))))...))))..)((((.............-.))))...)))). ( -38.74) >DroVir_CAF1 8911 107 + 1 UUGCAUUUGGUCUGUGUGCGUGCUCAGCGGAAGUAAAUAAAAGCUGAUGUCCUGUCAAGGCAGACCAUCAUUCGAGUGCCUGCGUUCAACAAGUUC-AAUUCUAUCA--G (((.(((((...((..((((..(((.((....))..........(((((..((((....))))..)))))...)))..)..)))..)).))))).)-))........--. ( -28.40) >DroGri_CAF1 10127 109 + 1 UGGCACUAUGUCUGUGUGCGUUCUCAGCGGAAGUAAAUAAAAGCUGAUGUCCUGCCACAGCAGAGCAUCAUUCGAGUGCCUGCGUUCAACAAGUUC-AAUUCAAGGAACA .((((((..........((.......((....))........))(((((..((((....))))..)))))....))))))............((((-........)))). ( -30.06) >DroSec_CAF1 7961 109 + 1 CACCGCGGAAUCUGCGCUCGUCCUCGGCGGAAGUAAAUAAAAGCGGAUGUCCUGGCUGAACAGGACAUCAUUCGAGUGCUUGCGUUCAACAAGUCC-AGCAAACAGAGCA ....((((...))))(((((..((((((....))...........(((((((((......)))))))))...))))..)((((.............-.))))...)))). ( -38.74) >DroWil_CAF1 8374 107 + 1 UUGCCAUUUGUCUGUGCGCGUCCACAACGGAAGUGAAUAAAAGCUGAUGUCCUGUCGUUAGAGGACAUCAUUCGAGUGCUUGCGUUCAACAAGUU---ACAGUCAGUGCU (((..((((((.((.(((((..(((.((....))..........(((((((((........))))))))).....)))..))))).)))))))).---.)))........ ( -32.00) >DroAna_CAF1 7931 110 + 1 CAUCGUGUCUUCUGGGCUCUUCCUCGACGGAAGUAAAUAAAAGCGGAUGUCCUGUGCAGGCAGGACAUCAUUCAAGUUCUUGCGUUCAACAAGUUCAAGUAACUAGUGCA ......(((.....))).(((((.....))))).........(((((((((((((....)))))))))).....((((((((..((.....))..)))).))))..))). ( -29.90) >consensus CAGCGCGUCGUCUGUGCGCGUCCUCAGCGGAAGUAAAUAAAAGCGGAUGUCCUGUCAAGACAGGACAUCAUUCGAGUGCUUGCGUUCAACAAGUUC_AACAAACAGAGCA ..............(((..(..(((.((....))...........(((((((((......)))))))))....)))..)..))).......................... (-20.44 = -21.05 + 0.61)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:48:16 2006