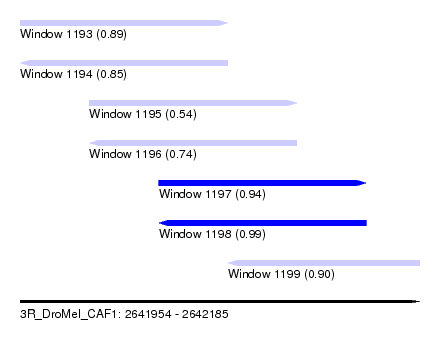
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 2,641,954 – 2,642,185 |
| Length | 231 |
| Max. P | 0.992924 |

| Location | 2,641,954 – 2,642,074 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -53.07 |
| Consensus MFE | -49.63 |
| Energy contribution | -50.30 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.891584 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2641954 120 + 27905053 AUCAGGUCGCAGCCUCUUGCAGGAUAAUGGCCGCAUUGCUGCUGCGACGCCAGCAGAAGCAGCGGCGGGCGAUGGCCGCAUUUUCUUGUUUGGUCUGUGAGCGGAACGCUCUACGUUCCG ....(.((((((((....((((((.(((((((((..((((.((((.......)))).))))))))).(((....))).)))).))))))..)).)))))).)((((((.....)))))). ( -57.60) >DroSim_CAF1 30371 120 + 1 AUCAGGUCGCAGCCUCCUGCAGGAUAAUGGCCGCAUUGCUGCUGCGACGCCAGCAGAAGCAGCGGCGGGCGAUGGCCGCAUUUUCUUGUUUGGUCUGUGAGCAGAACGCUCUACGUUCCG ....(.((((((((....((((((.(((((((((..((((.((((.......)))).))))))))).(((....))).)))).))))))..)).)))))).).(((((.....))))).. ( -53.30) >DroEre_CAF1 32829 120 + 1 AUCAGGUCGCAGCCUCCUGCAGGAUAAUGGCCGCACUGCUGCUGCGACGCCAGCUGAAGCAGCGGCGGGCGAUGGCCGCAUUUUCUUUUUUGGUCUGUGAGCAGAACGCUCUACGUUCCG ....(.(((((((((((....)))....((((((.((((((((((..((.....))..))))))))))))...))))..............)).)))))).).(((((.....))))).. ( -48.30) >consensus AUCAGGUCGCAGCCUCCUGCAGGAUAAUGGCCGCAUUGCUGCUGCGACGCCAGCAGAAGCAGCGGCGGGCGAUGGCCGCAUUUUCUUGUUUGGUCUGUGAGCAGAACGCUCUACGUUCCG ....(.((((((((....((((((.(((((((((..((((.((((.......)))).))))))))).(((....))).)))).))))))..)).)))))).).(((((.....))))).. (-49.63 = -50.30 + 0.67)

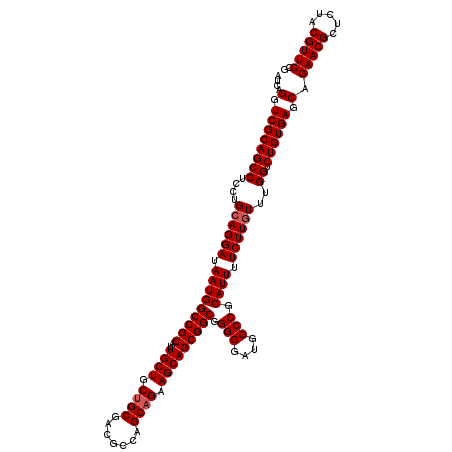
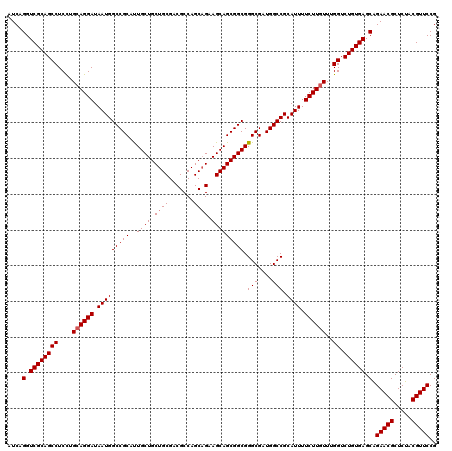
| Location | 2,641,954 – 2,642,074 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -46.80 |
| Consensus MFE | -46.52 |
| Energy contribution | -46.63 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.849949 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2641954 120 - 27905053 CGGAACGUAGAGCGUUCCGCUCACAGACCAAACAAGAAAAUGCGGCCAUCGCCCGCCGCUGCUUCUGCUGGCGUCGCAGCAGCAAUGCGGCCAUUAUCCUGCAAGAGGCUGCGACCUGAU (((((((.....)))))))....(((.(((....((((...(((((........)))))...))))..))).((((((((..(..(((((........))))).)..))))))))))).. ( -48.10) >DroSim_CAF1 30371 120 - 1 CGGAACGUAGAGCGUUCUGCUCACAGACCAAACAAGAAAAUGCGGCCAUCGCCCGCCGCUGCUUCUGCUGGCGUCGCAGCAGCAAUGCGGCCAUUAUCCUGCAGGAGGCUGCGACCUGAU (((((((.....)))))))....(((.(((....((((...(((((........)))))...))))..))).((((((((.....(((((........)))))....))))))))))).. ( -47.00) >DroEre_CAF1 32829 120 - 1 CGGAACGUAGAGCGUUCUGCUCACAGACCAAAAAAGAAAAUGCGGCCAUCGCCCGCCGCUGCUUCAGCUGGCGUCGCAGCAGCAGUGCGGCCAUUAUCCUGCAGGAGGCUGCGACCUGAU (((((((.....)))))))....(((...............(.(((....))))(((((((...)))).)))((((((((..(..(((((........))))).)..))))))))))).. ( -45.30) >consensus CGGAACGUAGAGCGUUCUGCUCACAGACCAAACAAGAAAAUGCGGCCAUCGCCCGCCGCUGCUUCUGCUGGCGUCGCAGCAGCAAUGCGGCCAUUAUCCUGCAGGAGGCUGCGACCUGAU (((((((.....)))))))....(((.(((....((((...(((((........)))))...))))..))).((((((((..(..(((((........))))).)..))))))))))).. (-46.52 = -46.63 + 0.11)

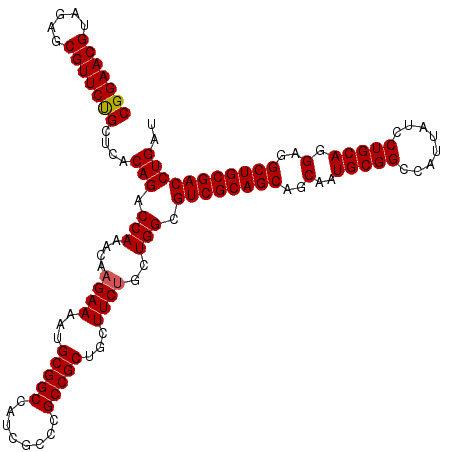
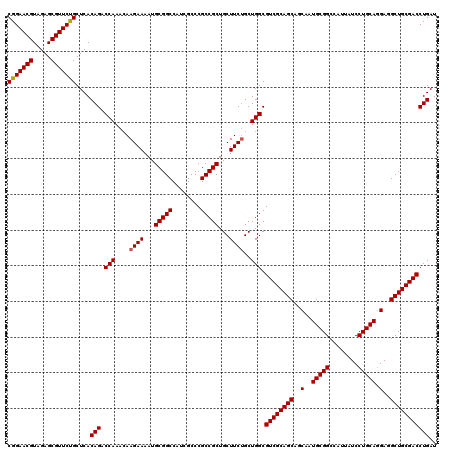
| Location | 2,641,994 – 2,642,114 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.78 |
| Mean single sequence MFE | -46.77 |
| Consensus MFE | -45.17 |
| Energy contribution | -45.50 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.535546 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2641994 120 + 27905053 GCUGCGACGCCAGCAGAAGCAGCGGCGGGCGAUGGCCGCAUUUUCUUGUUUGGUCUGUGAGCGGAACGCUCUACGUUCCGCCGCGCUCGGCGUCAAAUUUCAAACUGAAACUGAAGUGAG .....((((((((((((((..(((((.(....).)))))..)))))..........(((.((((((((.....)))))))))))))).))))))..((((((.........))))))... ( -48.90) >DroSim_CAF1 30411 120 + 1 GCUGCGACGCCAGCAGAAGCAGCGGCGGGCGAUGGCCGCAUUUUCUUGUUUGGUCUGUGAGCAGAACGCUCUACGUUCCGCCGCGCUCGGCGUCAAAUUUCAAACUAAAACUGAAGUGAG .....((((((.((....)).((((((((((..(((((((......)))..))))...((((.....))))..)).))))))))....))))))..((((((.........))))))... ( -46.50) >DroEre_CAF1 32869 120 + 1 GCUGCGACGCCAGCUGAAGCAGCGGCGGGCGAUGGCCGCAUUUUCUUUUUUGGUCUGUGAGCAGAACGCUCUACGUUCCGCCGCGCUCGGCGUCAAAUUUCAAACUGAAACUGAAGUGAG .....((((((.((....)).((((((((((..(((((.(........).)))))...((((.....))))..)).))))))))....))))))..((((((.........))))))... ( -44.90) >consensus GCUGCGACGCCAGCAGAAGCAGCGGCGGGCGAUGGCCGCAUUUUCUUGUUUGGUCUGUGAGCAGAACGCUCUACGUUCCGCCGCGCUCGGCGUCAAAUUUCAAACUGAAACUGAAGUGAG .....((((((.((....)).((((((((((..(((((.((......)).)))))...((((.....))))..)).))))))))....))))))..((((((.........))))))... (-45.17 = -45.50 + 0.33)

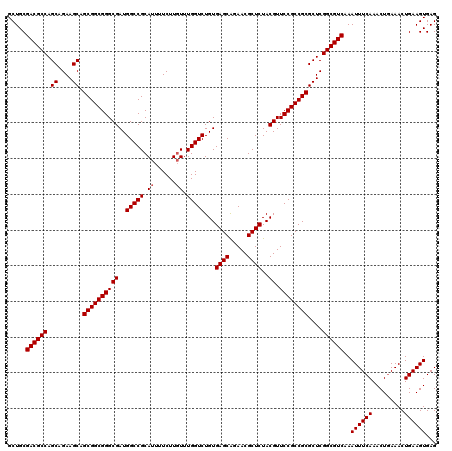
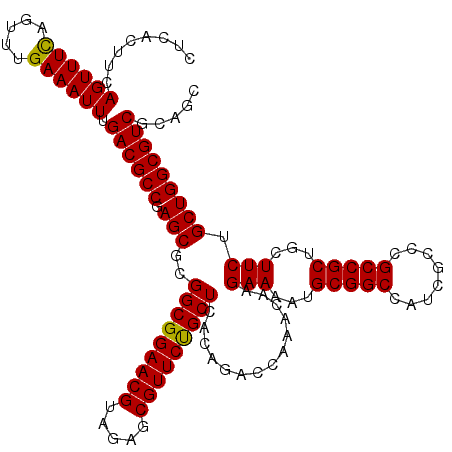
| Location | 2,641,994 – 2,642,114 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.78 |
| Mean single sequence MFE | -44.03 |
| Consensus MFE | -43.11 |
| Energy contribution | -42.67 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.742401 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2641994 120 - 27905053 CUCACUUCAGUUUCAGUUUGAAAUUUGACGCCGAGCGCGGCGGAACGUAGAGCGUUCCGCUCACAGACCAAACAAGAAAAUGCGGCCAUCGCCCGCCGCUGCUUCUGCUGGCGUCGCAGC ........((((((.....)))))).((((((.(((..(((((((((.....))))))))).............((((...(((((........)))))...)))))))))))))..... ( -45.90) >DroSim_CAF1 30411 120 - 1 CUCACUUCAGUUUUAGUUUGAAAUUUGACGCCGAGCGCGGCGGAACGUAGAGCGUUCUGCUCACAGACCAAACAAGAAAAUGCGGCCAUCGCCCGCCGCUGCUUCUGCUGGCGUCGCAGC .....(((((.......)))))....((((((.(((..(((((((((.....))))))))).............((((...(((((........)))))...)))))))))))))..... ( -43.00) >DroEre_CAF1 32869 120 - 1 CUCACUUCAGUUUCAGUUUGAAAUUUGACGCCGAGCGCGGCGGAACGUAGAGCGUUCUGCUCACAGACCAAAAAAGAAAAUGCGGCCAUCGCCCGCCGCUGCUUCAGCUGGCGUCGCAGC ........((((((.....)))))).((((((.(((..(((((((((.....)))))))))..............(((...(((((........)))))...))).)))))))))..... ( -43.20) >consensus CUCACUUCAGUUUCAGUUUGAAAUUUGACGCCGAGCGCGGCGGAACGUAGAGCGUUCUGCUCACAGACCAAACAAGAAAAUGCGGCCAUCGCCCGCCGCUGCUUCUGCUGGCGUCGCAGC ........((((((.....)))))).((((((.(((..(((((((((.....)))))))))..............(((...(((((........)))))...))).)))))))))..... (-43.11 = -42.67 + -0.44)

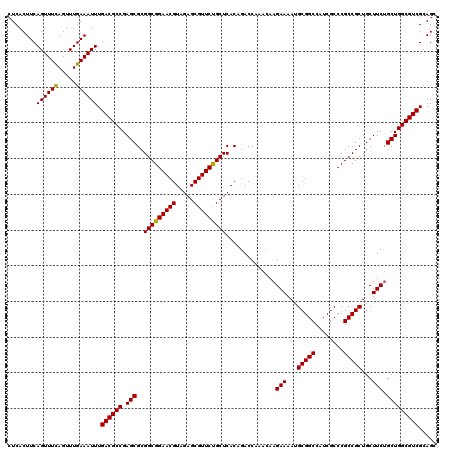
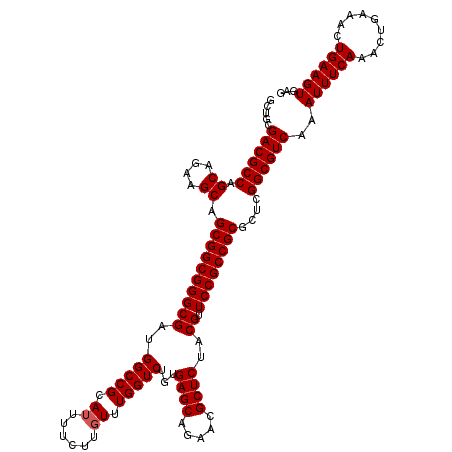
| Location | 2,642,034 – 2,642,154 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -44.00 |
| Consensus MFE | -41.70 |
| Energy contribution | -42.03 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.944983 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2642034 120 + 27905053 UUUUCUUGUUUGGUCUGUGAGCGGAACGCUCUACGUUCCGCCGCGCUCGGCGUCAAAUUUCAAACUGAAACUGAAGUGAGCUCAGCUCACCAUUUCAGCGCAGUCGACGUCCGCCGAGCG ................(((.((((((((.....)))))))))))((((((((....((.((..((((...(((((((((((...))))))...)))))..)))).)).)).)))))))). ( -49.50) >DroSim_CAF1 30451 120 + 1 UUUUCUUGUUUGGUCUGUGAGCAGAACGCUCUACGUUCCGCCGCGCUCGGCGUCAAAUUUCAAACUAAAACUGAAGUGAGCUCAGCUCACCAUUUCAGCGCAGUCGACGUCCGCCGAGCG ................(((.((.(((((.....))))).)))))((((((((....((.((..(((....(((((((((((...))))))...)))))...))).)).)).)))))))). ( -39.10) >DroEre_CAF1 32909 120 + 1 UUUUCUUUUUUGGUCUGUGAGCAGAACGCUCUACGUUCCGCCGCGCUCGGCGUCAAAUUUCAAACUGAAACUGAAGUGAGCUCAGCUCACCAUUUCAGCGCAGUCGACGUCCGCCGAGCG ................(((.((.(((((.....))))).)))))((((((((....((.((..((((...(((((((((((...))))))...)))))..)))).)).)).)))))))). ( -43.40) >consensus UUUUCUUGUUUGGUCUGUGAGCAGAACGCUCUACGUUCCGCCGCGCUCGGCGUCAAAUUUCAAACUGAAACUGAAGUGAGCUCAGCUCACCAUUUCAGCGCAGUCGACGUCCGCCGAGCG ................(((.((.(((((.....))))).)))))((((((((....((.((..((((...(((((((((((...))))))...)))))..)))).)).)).)))))))). (-41.70 = -42.03 + 0.33)

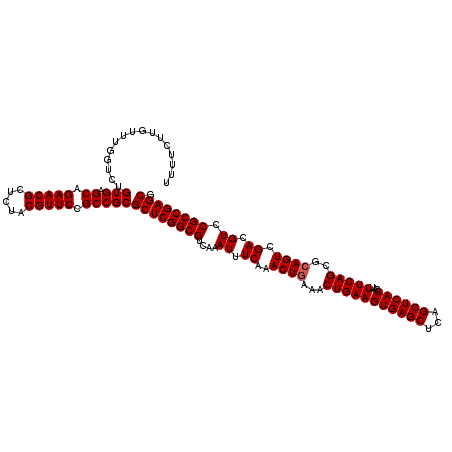
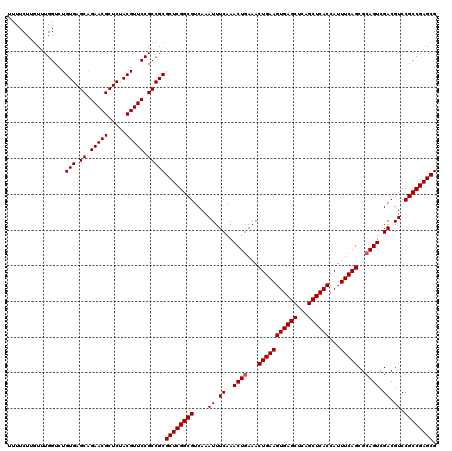
| Location | 2,642,034 – 2,642,154 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -51.33 |
| Consensus MFE | -51.77 |
| Energy contribution | -51.33 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.99 |
| Structure conservation index | 1.01 |
| SVM decision value | 2.36 |
| SVM RNA-class probability | 0.992924 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2642034 120 - 27905053 CGCUCGGCGGACGUCGACUGCGCUGAAAUGGUGAGCUGAGCUCACUUCAGUUUCAGUUUGAAAUUUGACGCCGAGCGCGGCGGAACGUAGAGCGUUCCGCUCACAGACCAAACAAGAAAA (((((((((....(((((((.(((((...(((((((...))))))))))))..))).)))).......))))))))).(((((((((.....)))))))))................... ( -53.40) >DroSim_CAF1 30451 120 - 1 CGCUCGGCGGACGUCGACUGCGCUGAAAUGGUGAGCUGAGCUCACUUCAGUUUUAGUUUGAAAUUUGACGCCGAGCGCGGCGGAACGUAGAGCGUUCUGCUCACAGACCAAACAAGAAAA (((((((((....(((((((.(((((...(((((((...))))))))))))..))).)))).......))))))))).(((((((((.....)))))))))................... ( -49.40) >DroEre_CAF1 32909 120 - 1 CGCUCGGCGGACGUCGACUGCGCUGAAAUGGUGAGCUGAGCUCACUUCAGUUUCAGUUUGAAAUUUGACGCCGAGCGCGGCGGAACGUAGAGCGUUCUGCUCACAGACCAAAAAAGAAAA (((((((((....(((((((.(((((...(((((((...))))))))))))..))).)))).......))))))))).(((((((((.....)))))))))................... ( -51.20) >consensus CGCUCGGCGGACGUCGACUGCGCUGAAAUGGUGAGCUGAGCUCACUUCAGUUUCAGUUUGAAAUUUGACGCCGAGCGCGGCGGAACGUAGAGCGUUCUGCUCACAGACCAAACAAGAAAA (((((((((....(((((((.(((((...(((((((...))))))))))))..))).)))).......))))))))).(((((((((.....)))))))))................... (-51.77 = -51.33 + -0.44)

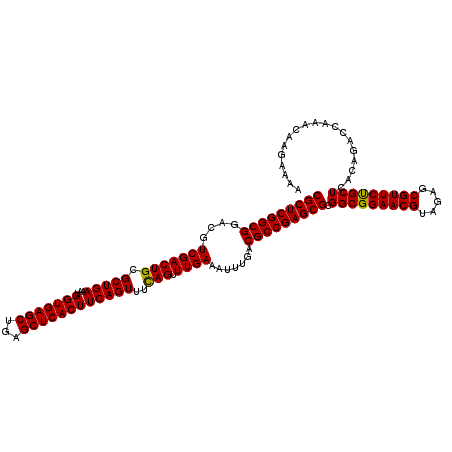
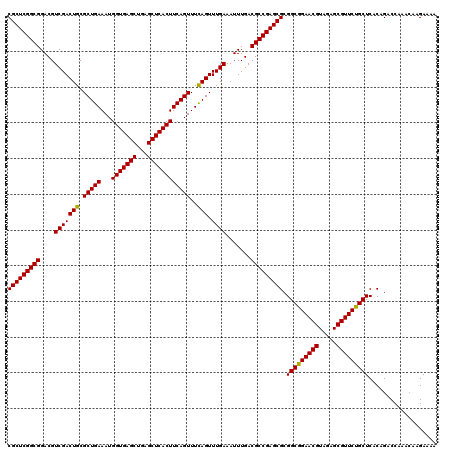
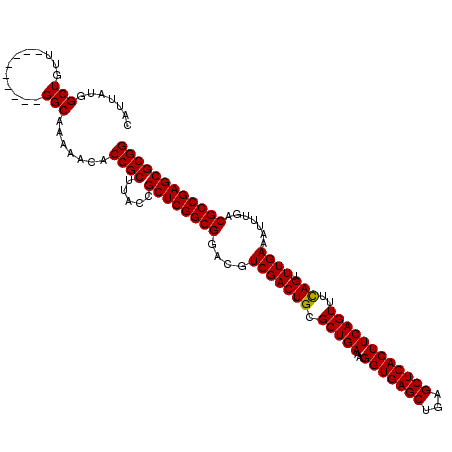
| Location | 2,642,074 – 2,642,185 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.30 |
| Mean single sequence MFE | -43.07 |
| Consensus MFE | -40.82 |
| Energy contribution | -40.60 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.898320 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2642074 111 - 27905053 CAUUAUGGCUGUU---------GGCAAAAACACCGCUUACCGCUCGGCGGACGUCGACUGCGCUGAAAUGGUGAGCUGAGCUCACUUCAGUUUCAGUUUGAAAUUUGACGCCGAGCGCGG ......((.((((---------......)))))).....((((((((((....(((((((.(((((...(((((((...))))))))))))..))).)))).......))))))))).). ( -43.30) >DroSim_CAF1 30491 111 - 1 CAUUAUGGCUGUU---------GGCAAAAACACCGCUUACCGCUCGGCGGACGUCGACUGCGCUGAAAUGGUGAGCUGAGCUCACUUCAGUUUUAGUUUGAAAUUUGACGCCGAGCGCGG ......((.((((---------......)))))).....((((((((((....(((((((.(((((...(((((((...))))))))))))..))).)))).......))))))))).). ( -41.50) >DroEre_CAF1 32949 120 - 1 CAUUAUGGCUGUUAUCAACGUGGGCAAAAACACCGCUUACCGCUCGGCGGACGUCGACUGCGCUGAAAUGGUGAGCUGAGCUCACUUCAGUUUCAGUUUGAAAUUUGACGCCGAGCGCGG .......((..........((((((.........)))))).((((((((....(((((((.(((((...(((((((...))))))))))))..))).)))).......)))))))))).. ( -44.40) >consensus CAUUAUGGCUGUU_________GGCAAAAACACCGCUUACCGCUCGGCGGACGUCGACUGCGCUGAAAUGGUGAGCUGAGCUCACUUCAGUUUCAGUUUGAAAUUUGACGCCGAGCGCGG .......(((............))).......((((.....((((((((....(((((((.(((((...(((((((...))))))))))))..))).)))).......)))))))))))) (-40.82 = -40.60 + -0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:51:13 2006