
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 20,072,547 – 20,072,690 |
| Length | 143 |
| Max. P | 0.994833 |

| Location | 20,072,547 – 20,072,651 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 76.08 |
| Mean single sequence MFE | -39.42 |
| Consensus MFE | -25.18 |
| Energy contribution | -27.38 |
| Covariance contribution | 2.20 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.658111 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
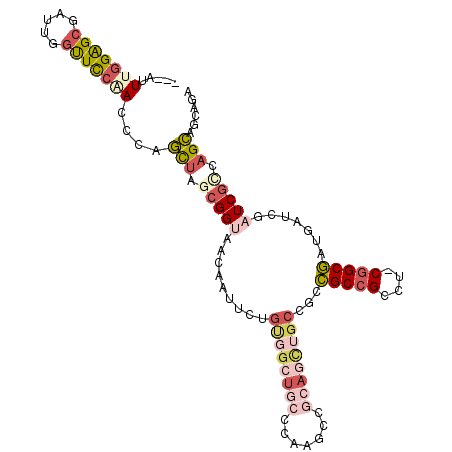
>3R_DroMel_CAF1 20072547 104 + 27905053 ---AUUUGGAGCGAUUGGUUCCAACCCAGCUAGCGGUAACAAUUCUGCGGCUGCCCAAGCCGCAGCUGCCGCCGCCGCCU-CGGCGAUGAUCGAUCGUCAGCAGCAGA ---..(((((((.....)))))))....(((.((((((......((((((((.....)))))))).)))))).((((...-))))(((((....))))))))...... ( -43.40) >DroPse_CAF1 16664 89 + 1 CAAUUUCGGGGCCAUCGGCUCGAAU---GCUGGCGGAAGCAGU---------------GCAGCAGCAGCUGCGGCCGCUG-CAGCCAUGAUCGAUCGCCAGCAGCAGA ....((((((.(....).))))))(---((((((((..(((((---------------(((((....)))))....))))-)..))..((....)))))))))..... ( -38.00) >DroSec_CAF1 14733 105 + 1 ---AUUUGGAGGGAUUGGUUUCGACCCCCCUACGGGUAACAAUGCUGUUGAUCACAAAACUGAACUUGCCAACGCCGUCUGCGGCGAUGAUCGAUCGUCAGGAGCAGU ---(((((.((((...(((....))).)))).))))).....((((.(((((((......)))......((.(((((....))))).))........)))).)))).. ( -29.10) >DroSim_CAF1 15964 104 + 1 ---AUUUGGAGCGAUUGGUUCCAACCCAGCUAGCGGUAACAAUUCUGCGGCUGCCCAAGCCGCAGCUGCCGCCGCCGCCU-CGGCGAUGAUCGAUCGGCAGCAGCAGA ---..(((((((.....)))))))....(((..............(((((((.....)))))))((((((((((......-))))(((.....))))))))))))... ( -44.60) >DroEre_CAF1 17356 104 + 1 ---AUUUGGAGCGAUUGGUUCCAACCCAGUUAGCGGUAACAAUUCGGUGGCUGCCCAGGCCGCAGCUGCUGCCGCCGCCU-CGGCGAUGAUCGAUCGCCAGCAGCAGA ---..(((((((.....)))))))....(((.(((((.......(((..(((((.......)))))..)))..)))))..-.((((((.....)))))))))...... ( -39.50) >DroYak_CAF1 15195 104 + 1 ---AUUCGGAGCUAUUGGUUCCAACCCAGUAAGCGGCAACAAUUCGGUGGCUGCCCAGGCCGCAGCUGCUGCCGCCGCAU-CGGCGAUGAUCGAUCGCCAGCAGCAGA ---...((((((((((((.......))))).)))(....)..))))((((((.....)))))).((((((((....))..-.((((((.....))))))))))))... ( -41.90) >consensus ___AUUUGGAGCGAUUGGUUCCAACCCAGCUAGCGGUAACAAUUCUGUGGCUGCCCAAGCCGCAGCUGCCGCCGCCGCCU_CGGCGAUGAUCGAUCGCCAGCAGCAGA .....(((((((.....)))))))....(((.(((((.........((((((((.......))))))))...(((((....))))).......))))).)))...... (-25.18 = -27.38 + 2.20)


| Location | 20,072,584 – 20,072,690 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 75.60 |
| Mean single sequence MFE | -40.62 |
| Consensus MFE | -22.59 |
| Energy contribution | -25.82 |
| Covariance contribution | 3.22 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.86 |
| Structure conservation index | 0.56 |
| SVM decision value | 2.52 |
| SVM RNA-class probability | 0.994833 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
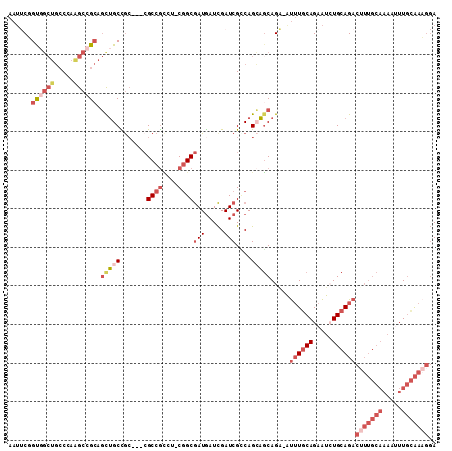
>3R_DroMel_CAF1 20072584 106 + 27905053 AAUUCUGCGGCUGCCCAAGCCGCAGCUGCCGC---CGCCGCCU-CGGCGAUGAUCGAUCGUCAGCAGCAGA-AUUUGCAGAAUCUGCAGACUUUGCAAAAUUUGCAAAGGA ..((((((((((.....)))))).(((((.((---((......-))))(((((....))))).))))))))-)(((((((...)))))))(((((((.....))))))).. ( -47.80) >DroSec_CAF1 14770 108 + 1 AAUGCUGUUGAUCACAAAACUGAACUUGCCAA---CGCCGUCUGCGGCGAUGAUCGAUCGUCAGGAGCAGUUGUUUGCAGAAUAUGCAGACUUUGCAAAAUUUGCAAAGGA ..((((.(((((((......)))......((.---(((((....))))).))........)))).))))....((((((.....))))))(((((((.....))))))).. ( -33.00) >DroSim_CAF1 16001 106 + 1 AAUUCUGCGGCUGCCCAAGCCGCAGCUGCCGC---CGCCGCCU-CGGCGAUGAUCGAUCGGCAGCAGCAGA-AUUUACAGAAUAUGCAGACUUUGCAAAAUUUGCAAAGGA ((((((((((((.....)))))).(((((.((---(((((...-))))(((.....)))))).))))))))-)))...............(((((((.....))))))).. ( -45.60) >DroEre_CAF1 17393 106 + 1 AAUUCGGUGGCUGCCCAGGCCGCAGCUGCUGC---CGCCGCCU-CGGCGAUGAUCGAUCGCCAGCAGCAGA-AUUUGCAGAAUCUGCAGACUUUGCAAAAUUUGCAAAGGA ..(((....(((((..((((.((.((....))---.)).))))-.((((((.....)))))).))))).))-)(((((((...)))))))(((((((.....))))))).. ( -45.00) >DroYak_CAF1 15232 106 + 1 AAUUCGGUGGCUGCCCAGGCCGCAGCUGCUGC---CGCCGCAU-CGGCGAUGAUCGAUCGCCAGCAGCAGA-AUUUGCAGAAUCUGCAGACAUUGCAAAACUUGCAACGGA ....(((..(((((.......)))))..)))(---((..(((.-.((((((.....))))))....((((.-.(((((((...)))))))..))))......)))..))). ( -42.20) >DroMoj_CAF1 18837 96 + 1 ----CGGUGCCCGCCUUAGCAGCAGCGGCUGCAAUAGCCAAUG-CAGCAAUAAUUGAUCGGCAGGUUCAGG-ACAUGCAGCGUCUGCAGCGU-------CUUUAUGAC--A ----..(.(((.(((...((....)).((((((........))-))))...........))).))).)(((-((.(((((...)))))..))-------)))......--. ( -30.10) >consensus AAUUCGGUGGCUGCCCAAGCCGCAGCUGCCGC___CGCCGCCU_CGGCGAUGAUCGAUCGCCAGCAGCAGA_AUUUGCAGAAUCUGCAGACUUUGCAAAAUUUGCAAAGGA ......((((((.....)))))).(((((.......((((....))))(((.....)))....))))).....((((((.....))))))(((((((.....))))))).. (-22.59 = -25.82 + 3.22)


| Location | 20,072,584 – 20,072,690 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 75.60 |
| Mean single sequence MFE | -41.67 |
| Consensus MFE | -22.69 |
| Energy contribution | -24.70 |
| Covariance contribution | 2.01 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.85 |
| Structure conservation index | 0.54 |
| SVM decision value | 2.50 |
| SVM RNA-class probability | 0.994657 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
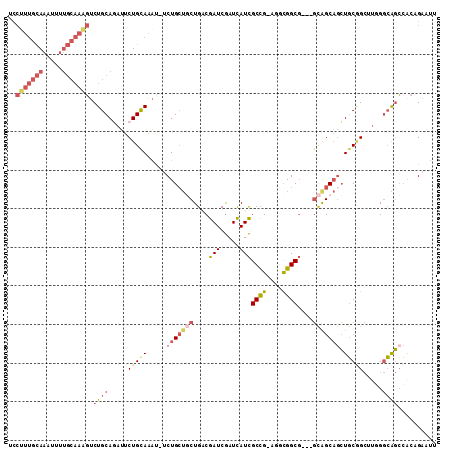
>3R_DroMel_CAF1 20072584 106 - 27905053 UCCUUUGCAAAUUUUGCAAAGUCUGCAGAUUCUGCAAAU-UCUGCUGCUGACGAUCGAUCAUCGCCG-AGGCGGCG---GCGGCAGCUGCGGCUUGGGCAGCCGCAGAAUU ..(((((((.....)))))))..(((((...)))))(((-(((((.(((((.((....)).))(((.-((((.(((---((....))))).)))).)))))).)))))))) ( -49.40) >DroSec_CAF1 14770 108 - 1 UCCUUUGCAAAUUUUGCAAAGUCUGCAUAUUCUGCAAACAACUGCUCCUGACGAUCGAUCAUCGCCGCAGACGGCG---UUGGCAAGUUCAGUUUUGUGAUCAACAGCAUU ..(((((((.....)))))))..((((.....))))...........(((..((((...((.(((((....)))))---.))(((((......)))))))))..))).... ( -29.00) >DroSim_CAF1 16001 106 - 1 UCCUUUGCAAAUUUUGCAAAGUCUGCAUAUUCUGUAAAU-UCUGCUGCUGCCGAUCGAUCAUCGCCG-AGGCGGCG---GCGGCAGCUGCGGCUUGGGCAGCCGCAGAAUU ..(((((((.....)))))))..((((.....))))...-.((((((((((((.(((........))-)..)))))---)))))))((((((((.....)))))))).... ( -49.50) >DroEre_CAF1 17393 106 - 1 UCCUUUGCAAAUUUUGCAAAGUCUGCAGAUUCUGCAAAU-UCUGCUGCUGGCGAUCGAUCAUCGCCG-AGGCGGCG---GCAGCAGCUGCGGCCUGGGCAGCCACCGAAUU ..(((((((.....)))))))..(((((...)))))(((-((.((((((((((((.....)))))).-((((.(((---((....))))).)))).))))))....))))) ( -48.20) >DroYak_CAF1 15232 106 - 1 UCCGUUGCAAGUUUUGCAAUGUCUGCAGAUUCUGCAAAU-UCUGCUGCUGGCGAUCGAUCAUCGCCG-AUGCGGCG---GCAGCAGCUGCGGCCUGGGCAGCCACCGAAUU ..(((((((.....)))))))..(((((...)))))(((-((((((((((.((((((........))-)).)).))---))))))(((((.......)))))....))))) ( -42.90) >DroMoj_CAF1 18837 96 - 1 U--GUCAUAAAG-------ACGCUGCAGACGCUGCAUGU-CCUGAACCUGCCGAUCAAUUAUUGCUG-CAUUGGCUAUUGCAGCCGCUGCUGCUAAGGCGGGCACCG---- .--........(-------(((.(((((...))))))))-).....((((((...........((((-((........)))))).((....))...)))))).....---- ( -31.00) >consensus UCCUUUGCAAAUUUUGCAAAGUCUGCAGAUUCUGCAAAU_UCUGCUGCUGACGAUCGAUCAUCGCCG_AGGCGGCG___GCAGCAGCUGCGGCUUGGGCAGCCACAGAAUU ..(((((((.....))))))).((((.....(((((.....(((((((....(((.....)))((((....))))....))))))).))))).....)))).......... (-22.69 = -24.70 + 2.01)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:47:47 2006