
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 20,066,006 – 20,066,148 |
| Length | 142 |
| Max. P | 0.963371 |

| Location | 20,066,006 – 20,066,117 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 76.40 |
| Mean single sequence MFE | -43.32 |
| Consensus MFE | -22.48 |
| Energy contribution | -23.87 |
| Covariance contribution | 1.39 |
| Combinations/Pair | 1.46 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.52 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.929054 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20066006 111 + 27905053 CCUCGGGCCGAGAUGAAGAACUUGGCCACGCUCUGCUCGGCCGCAGCAGCAGCUGCUGCUGUGGCAGCGGUUAACAAGGAUCAGGUUGAGAUUAGUUCGGAUCUUGAGAGC .....(((((((........)))))))..((((.(((..(((((((((((....))))))))))))))((((......))))...(..(((((......)))))..))))) ( -49.40) >DroVir_CAF1 12982 111 + 1 ACCCGGGCCGAAAUGAAGAAUCUGGCCACGCUCUGCUCUGCAGCCGCCAUGGCGGCUCAGGUGACAGCGGCAAAUAAGGAAAUCGAUGAAAUUGCUACCGAUAUCGAGAGU ..((.(((((............)))))..(((((((((((.((((((....)))))))))).).))).)))......))...(((((...((((....))))))))).... ( -36.00) >DroGri_CAF1 14247 111 + 1 ACGCGUGCAGAGAUGAAGAAUUUGGCCACGCUGUGCUCGGCAGCAGCCAUGGCGGCUGCCGUAACAGCAGCUAAUAAGGAGUCCGAAGAAGUUGCAACUGAUAACGAAAGU ..((((((..((((.....)))).).)))))(((...(((((((.((....)).)))))))..)))((((((.....((...)).....))))))........((....)) ( -38.60) >DroSec_CAF1 8171 111 + 1 CCUCCUGCCGAGAUGAAGAACUUGGCCACGCUGUGCUCGGCCGCAGCAGCAGCUGCUGCUGUGGCAGCGGUUAACAAGGAUCAGGUUGAGAUUAGUUCGGAUCUUGAGAGC ..((((((((((........))))))...(((((.....(((((((((((....))))))))))).))))).....)))).....(..(((((......)))))..).... ( -45.10) >DroEre_CAF1 10869 111 + 1 CCUAGGGCCGAGAUGAAAAAUUUGGCCACGCUCUGCUCGGCCGCAGCAGCAGCUGCUGCUGUGGCUGCGGUUAACAAGGAUCAGGAUGAGAUUAGUUCGGAACUUGAGAGC .....(((((((.(....).)))))))..(((((((((((((((((((((....))))))))))))).)))...((((..((.((((.......)))).)).))))))))) ( -50.50) >DroAna_CAF1 11710 111 + 1 CCUCGAUCCGAGAUGAAGAACCUGGCCACGCUCUGUUCAGCAGCAGCCGCAGCAGCGGCUGUGGCAGCCGCCAACAAGGAGCAGGACGAGAUCAGCACUGAUAUCGAAAGC .((((...))))........((((((((((((((....)).)))((((((....)))))))))))..((........))..)))).(((.((((....)))).)))..... ( -40.30) >consensus CCUCGGGCCGAGAUGAAGAACUUGGCCACGCUCUGCUCGGCAGCAGCAGCAGCUGCUGCUGUGGCAGCGGCUAACAAGGAUCAGGAUGAGAUUAGUACGGAUAUCGAGAGC .(((((((((((........))))))...((((((((((((.(((((....))))).)))).))))).)))................................)))))... (-22.48 = -23.87 + 1.39)



| Location | 20,066,037 – 20,066,148 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 74.06 |
| Mean single sequence MFE | -37.17 |
| Consensus MFE | -22.00 |
| Energy contribution | -21.20 |
| Covariance contribution | -0.80 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.825968 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20066037 111 + 27905053 UCUGCUCGGCCGCAGCAGCAGCUGCUGCUGUGGCAGCGGUUAACAAGGAUCAGGUUGAGAUUAGUUCGGAUCUUGAGAGCGA---AUGCGAGGAUGAUGCAGAAGGUGGUGCUG (((((((((((((((((((....))))))))))).((((((......))))...(..(((((......)))))..)..))..---.........))).)))))........... ( -41.60) >DroPse_CAF1 9795 96 + 1 UUUGUUCAGCAGCAGCAGCGGCCGCUGCUGUGGCAGCAGGCCACAGGGAUGAAGAUGAAAUCAGUACAGAUAUAUACACUGA------UGAGGAC------------GAUGCUA .((((((((((((.((....)).))))))(((((.....)))))...............((((((............)))))------)..))))------------))..... ( -29.80) >DroSec_CAF1 8202 105 + 1 UGUGCUCGGCCGCAGCAGCAGCUGCUGCUGUGGCAGCGGUUAACAAGGAUCAGGUUGAGAUUAGUUCGGAUCUUGAGAGCGA---AUGCGAGGAUGAC------GGUGGUGCUG .((.(((((((((((((((....))))))))))).((((((......))))...(..(((((......)))))..)..))..---...)))).))..(------((.....))) ( -40.40) >DroSim_CAF1 10156 105 + 1 UCUGCUCGGCCGCAGCAGCAGCUGCUGCUGUGGCAGCGGUUAACAAGGAUCAGGUUGAGAUUAGUUCGGAUCUUGAGAGCGA---AUGCGAGGAUGAC------GGUGGUGCUG (((.(((((((((((((((....))))))))))).((((((......))))...(..(((((......)))))..)..))..---...)))).).))(------((.....))) ( -39.30) >DroAna_CAF1 11741 108 + 1 UCUGUUCAGCAGCAGCCGCAGCAGCGGCUGUGGCAGCCGCCAACAAGGAGCAGGACGAGAUCAGCACUGAUAUCGAAAGCGACGGAGGCGAGGGCG------AAGGUGAUGCUG ......(((((((.((((((((....)))))))).))((((.....(..((....(((.((((....)))).)))...))..)...))))...((.------...))..))))) ( -39.30) >DroPer_CAF1 16388 96 + 1 UUUGUUCAGCAGCAGCAGCGGCCGCUGCUGUGGCAGCAGGCCACAAGGAUGAAGAUGACCUCAGUACAGAUCAAUACAGUGA------UGAGGAC------------GAUGCUA ((..(((((((((.((....)).))))))(((((.....)))))..)))..)).....(((((.(((...........))).------)))))..------------....... ( -32.60) >consensus UCUGCUCAGCAGCAGCAGCAGCUGCUGCUGUGGCAGCGGUCAACAAGGAUCAGGAUGAGAUCAGUACGGAUCUUGAGAGCGA___AUGCGAGGACG________GGUGAUGCUG ..(.((((((.((((((((....)))))))).)).............((((.......))))..........................)))).).................... (-22.00 = -21.20 + -0.80)
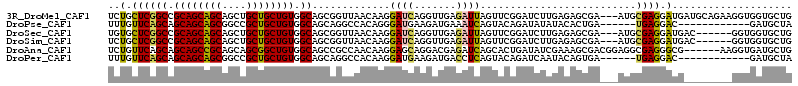
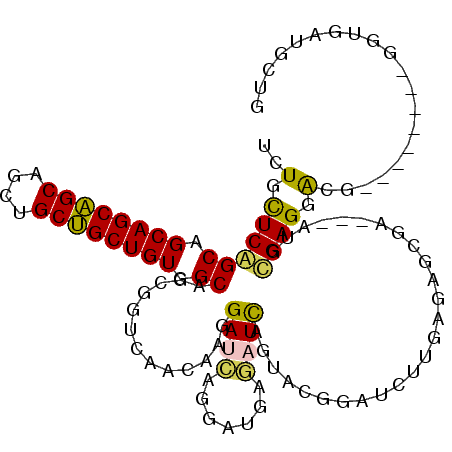

| Location | 20,066,037 – 20,066,148 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 74.06 |
| Mean single sequence MFE | -31.59 |
| Consensus MFE | -14.89 |
| Energy contribution | -14.70 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.47 |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.963371 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20066037 111 - 27905053 CAGCACCACCUUCUGCAUCAUCCUCGCAU---UCGCUCUCAAGAUCCGAACUAAUCUCAACCUGAUCCUUGUUAACCGCUGCCACAGCAGCAGCUGCUGCUGCGGCCGAGCAGA ...........(((((.((......((.(---(((.((....))..))))...(((.......)))...........)).(((.(((((((....))))))).))).))))))) ( -30.70) >DroPse_CAF1 9795 96 - 1 UAGCAUC------------GUCCUCA------UCAGUGUAUAUAUCUGUACUGAUUUCAUCUUCAUCCCUGUGGCCUGCUGCCACAGCAGCGGCCGCUGCUGCUGCUGAACAAA (((((.(------------......(------((((((((......)))))))))...............((((((.(((((....)))))))))))....).)))))...... ( -30.40) >DroSec_CAF1 8202 105 - 1 CAGCACCACC------GUCAUCCUCGCAU---UCGCUCUCAAGAUCCGAACUAAUCUCAACCUGAUCCUUGUUAACCGCUGCCACAGCAGCAGCUGCUGCUGCGGCCGAGCACA ..........------.............---..((((.(((((((.(.............).))).)))).........(((.(((((((....))))))).))).))))... ( -28.32) >DroSim_CAF1 10156 105 - 1 CAGCACCACC------GUCAUCCUCGCAU---UCGCUCUCAAGAUCCGAACUAAUCUCAACCUGAUCCUUGUUAACCGCUGCCACAGCAGCAGCUGCUGCUGCGGCCGAGCAGA ..........------.............---..((((.(((((((.(.............).))).)))).........(((.(((((((....))))))).))).))))... ( -28.32) >DroAna_CAF1 11741 108 - 1 CAGCAUCACCUU------CGCCCUCGCCUCCGUCGCUUUCGAUAUCAGUGCUGAUCUCGUCCUGCUCCUUGUUGGCGGCUGCCACAGCCGCUGCUGCGGCUGCUGCUGAACAGA (((((.......------.((..(((((..((..((...(((.((((....)))).)))....))....))..)))))..))..(((((((....))))))).)))))...... ( -38.40) >DroPer_CAF1 16388 96 - 1 UAGCAUC------------GUCCUCA------UCACUGUAUUGAUCUGUACUGAGGUCAUCUUCAUCCUUGUGGCCUGCUGCCACAGCAGCGGCCGCUGCUGCUGCUGAACAAA (((((.(------------(.(((((------.....((((......))))))))).)............((((((.(((((....)))))))))))....).)))))...... ( -33.40) >consensus CAGCACCACC________CAUCCUCGCAU___UCGCUCUCAAGAUCCGAACUAAUCUCAACCUGAUCCUUGUUAACCGCUGCCACAGCAGCAGCUGCUGCUGCGGCCGAACAGA .....................................................................((((....((.((...((((((....)))))))).))..)))).. (-14.89 = -14.70 + -0.19)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:47:40 2006