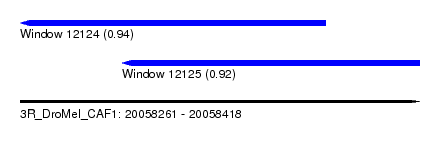
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 20,058,261 – 20,058,418 |
| Length | 157 |
| Max. P | 0.938404 |

| Location | 20,058,261 – 20,058,381 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.62 |
| Mean single sequence MFE | -43.50 |
| Consensus MFE | -28.03 |
| Energy contribution | -29.20 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.938404 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20058261 120 - 27905053 UGAUCCUGAUCCUCCUGCGCCUGAUCCACCGAGAGCAGCGCCAGCGUUGCUAUCGGCGGCAGCUGCGGCGGCGCCCAUCAGCAGAGUGGUUGUGGCGAAUUUCAAAUGCGCCGAAUCCUU ..............(((((.(((..((.((((.(((((((....))))))).)))).))))).)))))(((((((((.((((......))))))).(.....)....))))))....... ( -45.90) >DroGri_CAF1 4504 92 - 1 ----------------------------GCAGCAGCAUUACCAGCGUUGCCAUCUGUCGCAGAGGCAGCGGCGCCCAUCAACAAUGUUGUUGCGGCGAAUUUCAAAUGCGCCGAGUCCUU ----------------------------(((((((((((......((((((.((((...))))))))))((...))......)))))))))))((((...........))))........ ( -34.00) >DroEre_CAF1 3118 112 - 1 --------AUCCUCCUGCACCUGAUCCACCGGUAGCAGCGCCAGCGUUGCUAUCGGCGGCAGCUGCGGCGGCGCCCAUCAGCAGAGUGGUUGCGGCGAAUUUCAAAUGCGCCGAAUCCUU --------...((((((((.(((..((.((((((((((((....)))))))))))).))))).)))))..((........)).))).((...(((((...........)))))...)).. ( -47.50) >DroWil_CAF1 457 102 - 1 ------------------GGCGGCAGCGGUGGUAGCAGCGCCAGCGUUGCUAUCGACUGCAGCUGCAGCAGCGCCCAUUAGCAAUGUGGUGGCAGCAAAUUUCAAAUGUGCCGAAUCCUU ------------------..((((((((((((((((((((....)))))))))).))))).(((((.(((..((......))..)))....)))))............)))))....... ( -45.40) >DroYak_CAF1 946 112 - 1 --------AUCCUCCUGCACCUGAUCCACCGGUAGCAGCGCCAGCGUUGCUAUCGGCGGCAGCUGCGGCGGCGCCCAUCAGCAAAGUGGUUGCGGCGAAUUUCAAAUGCGCCGAAUCCUU --------......(((((.(((..((.((((((((((((....)))))))))))).))))).)))))((((((......((((.....))))...(.....)....))))))....... ( -47.40) >DroAna_CAF1 3798 104 - 1 -------------C---CUCCUCCUCCGCUGGUAGCAGCGCCAGCGUUGCUAUCCACAGCAGCAGCGGCGGCGCCCAUCAGCAGAGUGGUUGCGGCGAAUUUCAAAUGCGCCGAAUCCUU -------------.---......(((.((((((.((..((((.((((((((......)))))).))))))..))..)))))).))).((...(((((...........)))))...)).. ( -40.80) >consensus _____________C___CACCUGAUCCACCGGUAGCAGCGCCAGCGUUGCUAUCGGCGGCAGCUGCGGCGGCGCCCAUCAGCAAAGUGGUUGCGGCGAAUUUCAAAUGCGCCGAAUCCUU .............................(((((((((((....)))))))))))...((((((((....((........))...))))))))((((.((.....)).))))........ (-28.03 = -29.20 + 1.17)



| Location | 20,058,301 – 20,058,418 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 82.12 |
| Mean single sequence MFE | -44.82 |
| Consensus MFE | -38.42 |
| Energy contribution | -39.80 |
| Covariance contribution | 1.38 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.919032 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20058301 117 - 27905053 CCAUUCUGGCCCCGCCUACCGCUCCUACUGAUGAUGAUGAUCCUGAUCCUCCUGCGCCUGAUCCACCGAGAGCAGCGCCAGCGUUGCUAUCGGCGGCAGCUGCGGCGGCGCCCAUCA .......(((.(((((...................((.(((....))).))..(((.(((..((.((((.(((((((....))))))).)))).))))).)))))))).)))..... ( -44.30) >DroSec_CAF1 473 117 - 1 CCAUUCUGGCCCCGCCUACCGCUCCUACUGAUGAUGAUGAUCCUGAUCCUCCUGCGCCUGAUCCACCGGUAGCAGCGCCAGCGUUGCUAUCGGCGGCAGCUGCGGCGGCGCCCAUCA .......(((.(((((...................((.(((....))).))..(((.(((..((.((((((((((((....)))))))))))).))))).)))))))).)))..... ( -47.80) >DroSim_CAF1 929 117 - 1 CCAUUCUGGCCCCGCCUACCGCUCCUACUGAUGAUGAUGAUCCUGAUCCUCCUGCGCCUGAUCCACCGGUAGCAGCGCCAGCGUUGCUAUCGGCGGCAGCUGCGGCGGCGCCCAUCA .......(((.(((((...................((.(((....))).))..(((.(((..((.((((((((((((....)))))))))))).))))).)))))))).)))..... ( -47.80) >DroEre_CAF1 3158 108 - 1 CCAUUCUGGCCCCGCCUCCCGCUCCGCCAGAUGAUG---------AUCCUCCUGCACCUGAUCCACCGGUAGCAGCGCCAGCGUUGCUAUCGGCGGCAGCUGCGGCGGCGCCCAUCA ....((((((...((.....))...))))))(((((---------..(((.(((((.(((..((.((((((((((((....)))))))))))).))))).))))).)).)..))))) ( -50.10) >DroYak_CAF1 986 108 - 1 CCAUUCUGGCCCCGCCUCCCGCUCCACCAGAUGAUG---------AUCCUCCUGCACCUGAUCCACCGGUAGCAGCGCCAGCGUUGCUAUCGGCGGCAGCUGCGGCGGCGCCCAUCA .......(((.(((((.............(((....---------))).....(((.(((..((.((((((((((((....)))))))))))).))))).)))))))).)))..... ( -46.90) >DroAna_CAF1 3838 89 - 1 CCGGUUUGGACC-ACCGGCAGCACCU------------------------C---CUCCUCCUCCGCUGGUAGCAGCGCCAGCGUUGCUAUCCACAGCAGCAGCGGCGGCGCCCAUCA (((((.......-)))))..((.((.------------------------(---(.........((((((......))))))((((((......))))))...)).)).))...... ( -32.00) >consensus CCAUUCUGGCCCCGCCUACCGCUCCUACUGAUGAUG_________AUCCUCCUGCGCCUGAUCCACCGGUAGCAGCGCCAGCGUUGCUAUCGGCGGCAGCUGCGGCGGCGCCCAUCA .......(((.(((((.............((.(((.(......).))).))..(((.(((..((.((((((((((((....)))))))))))).))))).)))))))).)))..... (-38.42 = -39.80 + 1.38)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:47:38 2006