
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 20,057,200 – 20,057,305 |
| Length | 105 |
| Max. P | 0.987175 |

| Location | 20,057,200 – 20,057,305 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 91.27 |
| Mean single sequence MFE | -26.38 |
| Consensus MFE | -22.52 |
| Energy contribution | -23.52 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.953020 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20057200 105 + 27905053 UUGUUGAUGUGUUUCUGUGCCGCCGCCGUCACCGCCGUGACCCUACACACCUUUGAAAAACACAUCAACAAAGUUGUUAAAUUCACUUGUAGUGAAAUUUAUUUG ((((((((((((((..(.((....)))(((((....)))))................))))))))))))))......(((((((((.....))).)))))).... ( -27.30) >DroSec_CAF1 45669 105 + 1 UUGUUGAUGUGUUUCUGUGCCGCCGCCGUCACUGCCGUGCCCCUACACACCUUUGAAAAACACAUCAACAAAGUUGUUAAAUUCACUUGUAGUGAAAUUUAUUUU ((((((((((((((..(((.((....)).)))....(((......))).........))))))))))))))......(((((((((.....))).)))))).... ( -26.70) >DroSim_CAF1 47931 105 + 1 UUGUUGAUGUGUUUCUGUGCCGCCGCCGUCACCGCCGUGCCCCUACACUCCUUUGAAAAACACAUCAACAAAGUUGUUAAAUUCACUUGUAGUGAAAUUUAUUUG ((((((((((((((..(((.((....)).)))....(((......))).........))))))))))))))......(((((((((.....))).)))))).... ( -25.80) >DroYak_CAF1 48709 98 + 1 UUGUUGAUGUGUUUUUGUGCCGCCGCCG---CCGACGUGCCCC----CUCCCUUGAAAAACACAUCAACAAAGUUGUUAAAUUCACUUAGAGUGACAUUUACUUG ((((((((((((((((..((((.((...---.)).)).))...----(......)))))))))))))))))......(((((((((.....)))).))))).... ( -25.70) >consensus UUGUUGAUGUGUUUCUGUGCCGCCGCCGUCACCGCCGUGCCCCUACACACCUUUGAAAAACACAUCAACAAAGUUGUUAAAUUCACUUGUAGUGAAAUUUAUUUG ((((((((((((((..(((.((....)).)))....(((......))).........))))))))))))))......(((((((((.....)))).))))).... (-22.52 = -23.52 + 1.00)

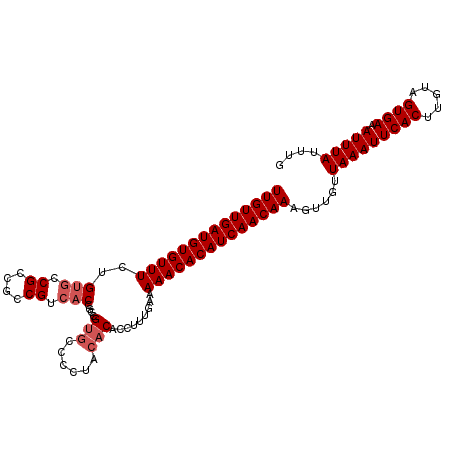

| Location | 20,057,200 – 20,057,305 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 91.27 |
| Mean single sequence MFE | -34.08 |
| Consensus MFE | -28.85 |
| Energy contribution | -29.91 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.65 |
| Structure conservation index | 0.85 |
| SVM decision value | 2.07 |
| SVM RNA-class probability | 0.987175 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20057200 105 - 27905053 CAAAUAAAUUUCACUACAAGUGAAUUUAACAACUUUGUUGAUGUGUUUUUCAAAGGUGUGUAGGGUCACGGCGGUGACGGCGGCGGCACAGAAACACAUCAACAA ....((((((.(((.....)))))))))......(((((((((((((((.......(((((.......((.((....)).))...)))))))))))))))))))) ( -34.81) >DroSec_CAF1 45669 105 - 1 AAAAUAAAUUUCACUACAAGUGAAUUUAACAACUUUGUUGAUGUGUUUUUCAAAGGUGUGUAGGGGCACGGCAGUGACGGCGGCGGCACAGAAACACAUCAACAA ....((((((.(((.....)))))))))......((((((((((((((((....(.(((((....))))).).(((.((....)).))))))))))))))))))) ( -33.80) >DroSim_CAF1 47931 105 - 1 CAAAUAAAUUUCACUACAAGUGAAUUUAACAACUUUGUUGAUGUGUUUUUCAAAGGAGUGUAGGGGCACGGCGGUGACGGCGGCGGCACAGAAACACAUCAACAA ....((((((.(((.....)))))))))......(((((((((((((((((....))((((....((.((.((....)).)))).)))).))))))))))))))) ( -37.20) >DroYak_CAF1 48709 98 - 1 CAAGUAAAUGUCACUCUAAGUGAAUUUAACAACUUUGUUGAUGUGUUUUUCAAGGGAG----GGGGCACGUCGG---CGGCGGCGGCACAAAAACACAUCAACAA ....(((((.((((.....)))))))))......(((((((((((((((((....)).----...((.(((((.---...)))))))...))))))))))))))) ( -30.50) >consensus CAAAUAAAUUUCACUACAAGUGAAUUUAACAACUUUGUUGAUGUGUUUUUCAAAGGAGUGUAGGGGCACGGCGGUGACGGCGGCGGCACAGAAACACAUCAACAA ....(((((.((((.....)))))))))......((((((((((((((((.......((((....((.((.((....)).)))).)))))))))))))))))))) (-28.85 = -29.91 + 1.06)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:47:36 2006