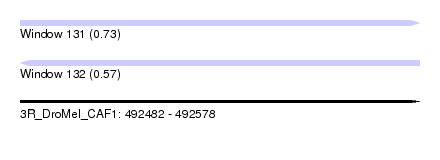
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 492,482 – 492,578 |
| Length | 96 |
| Max. P | 0.726945 |

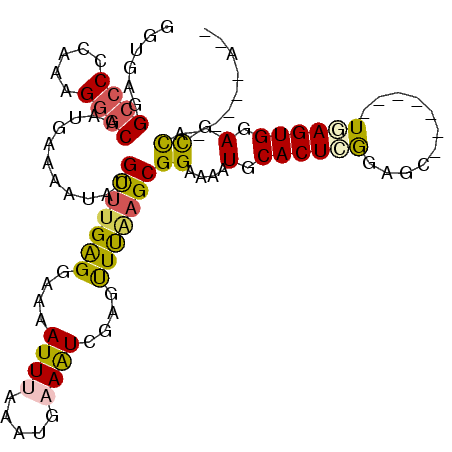
| Location | 492,482 – 492,578 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 79.96 |
| Mean single sequence MFE | -24.93 |
| Consensus MFE | -15.23 |
| Energy contribution | -14.85 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.726945 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 492482 96 + 27905053 GGUGAGGCCCCCAAAGGGCAAUGAAAUUUUGCGUGGGGAAAAUUUAAAUGAAGUGUAGUUUAAGCGGAAAAUGCACUCGGAGC-------UGAGUGGAGCCA-----A-- .....((((((((....((((.......)))).)))))....(((((((........))))))).......(.((((((....-------)))))).)))).-----.-- ( -25.50) >DroSec_CAF1 20534 96 + 1 GGUGAGGCCCCCAAAGGGCAAUGAAAAUUUGCUUGGGGAAAAUUUAAAUGAAGUGUAGUUUGAGCGGAAAAUGCACUCGGAGC-------UGAGUGGAGCCA-----A-- .....((((((((...(((((.......))))))))))....(((((((........))))))).......(.((((((....-------)))))).)))).-----.-- ( -26.40) >DroSim_CAF1 20026 96 + 1 GGUGAGGCCCCCAAAGGGCAAUGAAAAUUUGCUUGGGGAAAAUUUAAAUGAAGUGUAGUUUAAGCGGAAAAUGCACUCGGAGC-------UGAGUGGAGCCA-----A-- .....((((((((...(((((.......))))))))))....(((((((........))))))).......(.((((((....-------)))))).)))).-----.-- ( -26.30) >DroEre_CAF1 22606 100 + 1 UGUGAGGG-CCCAAAGGGCAAGGAAAAUAUGCUUGAGGACAAUU-A-AUGAAAUCGUGCUUAAGCGGAAAAUGCACUGGGAGCCAGCGGCUGAGUGGAGUCA-----A-- .((...((-(((...(((((.........((((((((.((.(((-.-....))).)).)))))))).....))).)).)).))).))((((......)))).-----.-- ( -27.84) >DroYak_CAF1 36949 95 + 1 GGUGAAGCCCCCAAAGGGCAACGAAAAUAAGUUGGAGGAAAAUU-AAAUGAAAUCGUAUUUAAGCGGAAAAUGCACUUGGAGC-------UAAGUGGAGUCA-----A-- ......((((.....))))(((........))).........((-(((((......)))))))...((...(.((((((....-------)))))).).)).-----.-- ( -16.10) >DroAna_CAF1 21538 110 + 1 CGAGAGGCCCCCAAAGGGCAAGGAAAAAAUGCUUGAGGAAAAUUUAAAUGAAAUCGUGUUCAAGCGGAAAAUGCACUCAGAGUUAGGAGUUAAGUGGAGUUGGUCUCUGG (.(((((((((((....(((.........((((((((.(..((((.....))))..).)))))))).....)))((((........))))....))).)..))))))).) ( -27.44) >consensus GGUGAGGCCCCCAAAGGGCAAUGAAAAUAUGCUUGAGGAAAAUUUAAAUGAAAUCGAGUUUAAGCGGAAAAUGCACUCGGAGC_______UGAGUGGAGCCA_____A__ ......((((.....))))...........(((((((....((((.....))))....)))))))((....(.((((((...........)))))).).))......... (-15.23 = -14.85 + -0.38)

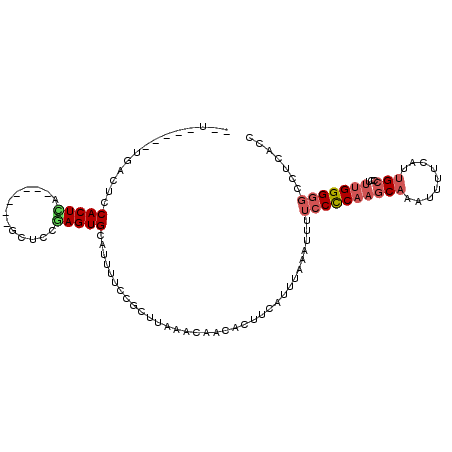
| Location | 492,482 – 492,578 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 79.96 |
| Mean single sequence MFE | -20.10 |
| Consensus MFE | -8.58 |
| Energy contribution | -9.22 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.43 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.573197 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 492482 96 - 27905053 --U-----UGGCUCCACUCA-------GCUCCGAGUGCAUUUUCCGCUUAAACUACACUUCAUUUAAAUUUUCCCCACGCAAAAUUUCAUUGCCCUUUGGGGGCCUCACC --.-----.(((..(((((.-------.....)))))...................................(((((.((((.......))))....))))))))..... ( -20.30) >DroSec_CAF1 20534 96 - 1 --U-----UGGCUCCACUCA-------GCUCCGAGUGCAUUUUCCGCUCAAACUACACUUCAUUUAAAUUUUCCCCAAGCAAAUUUUCAUUGCCCUUUGGGGGCCUCACC --.-----.(((..(((((.-------.....)))))...................................((((((((((.......))))...)))))))))..... ( -21.50) >DroSim_CAF1 20026 96 - 1 --U-----UGGCUCCACUCA-------GCUCCGAGUGCAUUUUCCGCUUAAACUACACUUCAUUUAAAUUUUCCCCAAGCAAAUUUUCAUUGCCCUUUGGGGGCCUCACC --.-----.(((..(((((.-------.....)))))...................................((((((((((.......))))...)))))))))..... ( -21.50) >DroEre_CAF1 22606 100 - 1 --U-----UGACUCCACUCAGCCGCUGGCUCCCAGUGCAUUUUCCGCUUAAGCACGAUUUCAU-U-AAUUGUCCUCAAGCAUAUUUUCCUUGCCCUUUGGG-CCCUCACA --.-----............((.(((((...))))))).......((((.((.((((((....-.-)))))).)).))))...........((((...)))-)....... ( -22.10) >DroYak_CAF1 36949 95 - 1 --U-----UGACUCCACUUA-------GCUCCAAGUGCAUUUUCCGCUUAAAUACGAUUUCAUUU-AAUUUUCCUCCAACUUAUUUUCGUUGCCCUUUGGGGGCUUCACC --.-----...........(-------((((((((((.......)))))................-..........((((........)))).......))))))..... ( -13.20) >DroAna_CAF1 21538 110 - 1 CCAGAGACCAACUCCACUUAACUCCUAACUCUGAGUGCAUUUUCCGCUUGAACACGAUUUCAUUUAAAUUUUCCUCAAGCAUUUUUUCCUUGCCCUUUGGGGGCCUCUCG ...((((.......((((((...........))))))........((((((....(((((.....)))))....))))))...........((((.....)))).)))). ( -22.00) >consensus __U_____UGACUCCACUCA_______GCUCCGAGUGCAUUUUCCGCUUAAACAACACUUCAUUUAAAUUUUCCCCAAGCAAAUUUUCAUUGCCCUUUGGGGGCCUCACC ..............(((((.............)))))..................................((((((((((.........)))...)))))))....... ( -8.58 = -9.22 + 0.64)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:32:26 2006