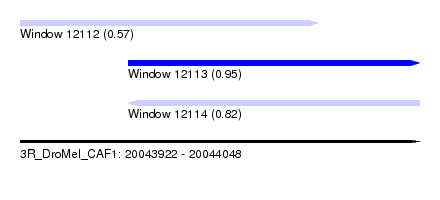
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 20,043,922 – 20,044,048 |
| Length | 126 |
| Max. P | 0.950991 |

| Location | 20,043,922 – 20,044,016 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 85.80 |
| Mean single sequence MFE | -41.97 |
| Consensus MFE | -33.73 |
| Energy contribution | -33.73 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.34 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.566835 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20043922 94 + 27905053 CGGCGCACUCAUUGAUCCGCUGCUGAUUAACCACUCCAUAGCCAACUGGUUGAGGUGGUUGGCCAGCCGACUG---AGCUGCCCAAUUGGGCGG-UGG .((((..(.....)...))))((((..(((((((..((..(((....)))))..)))))))..))))......---.(((((((....))))))-).. ( -36.80) >DroSec_CAF1 32386 94 + 1 CGGCGCACUCAUUGAUCCGCUGCUGGUUGACCACUCCAUAGCCAACAGGCUGAGGUGGUUGGCCAGCCGACUG---AGCUGCCCAAUUGGGCGG-UGG .((((..(.....)...))))((((((..((((((...(((((....))))).))))))..))))))......---.(((((((....))))))-).. ( -45.80) >DroSim_CAF1 32113 94 + 1 CGGCGCACUCAUUGAUCCGCUGCUGGUUGACCACUCCAUAGCCAACAGGCUGAGGUGGUUGGCCAGCCGACUG---AGCUGCCCAAUUGGGCGG-UGG .((((..(.....)...))))((((((..((((((...(((((....))))).))))))..))))))......---.(((((((....))))))-).. ( -45.80) >DroEre_CAF1 34786 94 + 1 CGGCGCACUCAUUGAUCCGCUGCUGAUUGACCACUCCAUAGCCAGCAGGCUGAGGUGGUUGGCCGGCCGACUG---AACUGCCCAAUUGGGUGG-UGA .((((..(.....)...))))((((.(..((((((...(((((....))))).))))))..).))))......---.((..(((....)))..)-).. ( -35.40) >DroYak_CAF1 35300 94 + 1 CGGCGCACUCAUUGAUCCGCUGCUGGUUGACCAUGCCAUAGCCAGCAGGCUGAGGUGGUUGGCUAGCCGACUG---AACUGCCCAAUUGGGCGG-UGG .((((..(.....)...))))((((((..(((..(((.(((((....))))).))))))..))))))......---.(((((((....))))))-).. ( -42.30) >DroAna_CAF1 34155 98 + 1 CGGCGCAUUCGUUGAUCCGUUGCUGGUUGACGACGCCGUAGCCACUGGGCUGUGGCGGCGGACCAGUCGCCGGCGAGGUGGUCCAGCUGGGGGGAUGG ((((((..((((..(((.......)))..))))((((((((((....))))))))))))((((((.(((....)))..)))))).))))......... ( -45.70) >consensus CGGCGCACUCAUUGAUCCGCUGCUGGUUGACCACUCCAUAGCCAACAGGCUGAGGUGGUUGGCCAGCCGACUG___AGCUGCCCAAUUGGGCGG_UGG .((((............))))((((((((((((..((.(((((....))))).))))))))))))))...........((((((....)))))).... (-33.73 = -33.73 + 0.01)


| Location | 20,043,956 – 20,044,048 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 80.42 |
| Mean single sequence MFE | -43.73 |
| Consensus MFE | -29.17 |
| Energy contribution | -30.32 |
| Covariance contribution | 1.14 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.87 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.950991 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20043956 92 + 27905053 UCCAUAGCCAACUGGUUGAGGUGGUUGGCCAGCCGACUG---AGCUGCCCAAUUGGGCGG-UGGUGCAGACGGCGGACCAUGAUGCAACCUGUACA--- ....(((((....)))))((((.(((((((.((((.(((---.(((((((....))))))-)....))).)))))).)))....)).)))).....--- ( -39.00) >DroSec_CAF1 32420 92 + 1 UCCAUAGCCAACAGGCUGAGGUGGUUGGCCAGCCGACUG---AGCUGCCCAAUUGGGCGG-UGGUGCAGACGGCCGACCAUGAUGCAGCCUGUACA--- ..........((((((((..((((((((((..(.((((.---.(((((((....))))))-)))).).)..))))))))))....))))))))...--- ( -47.30) >DroSim_CAF1 32147 92 + 1 UCCAUAGCCAACAGGCUGAGGUGGUUGGCCAGCCGACUG---AGCUGCCCAAUUGGGCGG-UGGUGCAGACGGCGGACCAUGAUGCAGCCUGUACA--- ..........((((((((..((.((.((((.((((.(((---.(((((((....))))))-)....))).)))))).)))).)).))))))))...--- ( -45.40) >DroEre_CAF1 34820 92 + 1 UCCAUAGCCAGCAGGCUGAGGUGGUUGGCCGGCCGACUG---AACUGCCCAAUUGGGUGG-UGAUGCCGACGGCGGACCAUGAUGCAGCCUGUACA--- ..........((((((((..((((((.((((..((.(..---.((..(((....)))..)-)...).)).)))).))))))....))))))))...--- ( -45.80) >DroYak_CAF1 35334 92 + 1 GCCAUAGCCAGCAGGCUGAGGUGGUUGGCUAGCCGACUG---AACUGCCCAAUUGGGCGG-UGGUGCAGAAGGUGGACCAUGAUGCAGCCUAUACA--- (((.(((((....))))).)))((((((....)))))).---.(((((((....))))))-)((((((..(((....)).)..)))).))......--- ( -38.40) >DroAna_CAF1 34189 92 + 1 GCCGUAGCCACUGGGCUGUGGCGGCGGACCAGUCGCCGGCGAGGUGGUCCAGCUGGGGGGAUGG-------GGCGCCGCAUGAUGCAGUCGCUGCAAGU ...(((((.((((.(..(((.(((((..(((.((.(((((..((....)).))))).))..)))-------..))))))))..).)))).))))).... ( -46.50) >consensus UCCAUAGCCAACAGGCUGAGGUGGUUGGCCAGCCGACUG___AGCUGCCCAAUUGGGCGG_UGGUGCAGACGGCGGACCAUGAUGCAGCCUGUACA___ ..........((((((((..((((((.....((((.(((.....((((((....))))))......))).)))).))))))....))))))))...... (-29.17 = -30.32 + 1.14)



| Location | 20,043,956 – 20,044,048 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 80.42 |
| Mean single sequence MFE | -35.30 |
| Consensus MFE | -23.42 |
| Energy contribution | -26.23 |
| Covariance contribution | 2.81 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.817411 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20043956 92 - 27905053 ---UGUACAGGUUGCAUCAUGGUCCGCCGUCUGCACCA-CCGCCCAAUUGGGCAGCU---CAGUCGGCUGGCCAACCACCUCAACCAGUUGGCUAUGGA ---......(((.(((..((((....)))).)))))).-((((((....))))....---.((((((((((.............))))))))))..)). ( -33.52) >DroSec_CAF1 32420 92 - 1 ---UGUACAGGCUGCAUCAUGGUCGGCCGUCUGCACCA-CCGCCCAAUUGGGCAGCU---CAGUCGGCUGGCCAACCACCUCAGCCUGUUGGCUAUGGA ---.((((((((((.....((((((((((.(((.....-(.((((....)))).)..---))).)))))))))).......))))))))..))...... ( -41.80) >DroSim_CAF1 32147 92 - 1 ---UGUACAGGCUGCAUCAUGGUCCGCCGUCUGCACCA-CCGCCCAAUUGGGCAGCU---CAGUCGGCUGGCCAACCACCUCAGCCUGUUGGCUAUGGA ---.((((((((((.....(((((.((((.(((.....-(.((((....)))).)..---))).)))).))))).......))))))))..))...... ( -37.90) >DroEre_CAF1 34820 92 - 1 ---UGUACAGGCUGCAUCAUGGUCCGCCGUCGGCAUCA-CCACCCAAUUGGGCAGUU---CAGUCGGCCGGCCAACCACCUCAGCCUGCUGGCUAUGGA ---.((.(((((((.....((((..(((((((((...(-(..(((....)))..)).---..))))).))))..))))...)))))))...))...... ( -35.30) >DroYak_CAF1 35334 92 - 1 ---UGUAUAGGCUGCAUCAUGGUCCACCUUCUGCACCA-CCGCCCAAUUGGGCAGUU---CAGUCGGCUAGCCAACCACCUCAGCCUGCUGGCUAUGGC ---......((.((((....((....))...)))))).-((((((....))))(((.---((((.((((((........)).)))).)))))))..)). ( -29.50) >DroAna_CAF1 34189 92 - 1 ACUUGCAGCGACUGCAUCAUGCGGCGCC-------CCAUCCCCCCAGCUGGACCACCUCGCCGGCGACUGGUCCGCCGCCACAGCCCAGUGGCUACGGC ...((..(((.((((.....))))))).-------.)).....((.((.((((((..(((....))).)))))))).(((((......)))))...)). ( -33.80) >consensus ___UGUACAGGCUGCAUCAUGGUCCGCCGUCUGCACCA_CCGCCCAAUUGGGCAGCU___CAGUCGGCUGGCCAACCACCUCAGCCUGUUGGCUAUGGA ....((.(((((((.....(((((.((((.(((........((((....)))).......))).)))).))))).......)))))))...))...... (-23.42 = -26.23 + 2.81)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:47:27 2006