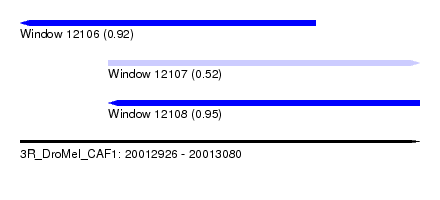
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 20,012,926 – 20,013,080 |
| Length | 154 |
| Max. P | 0.953828 |

| Location | 20,012,926 – 20,013,040 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.72 |
| Mean single sequence MFE | -32.77 |
| Consensus MFE | -22.87 |
| Energy contribution | -22.37 |
| Covariance contribution | -0.49 |
| Combinations/Pair | 1.54 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.924820 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
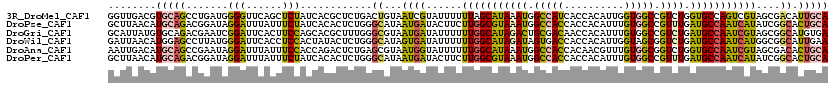
>3R_DroMel_CAF1 20012926 114 - 27905053 CUGACUGUAAUCGUAUUUUUUAGCAUAAAUGGCCAUCACCACAUUGGUGGCCGUCUGGUGCCAGUCGUAGCGACAUUGCACAACCUUUUUGUCGUCGUUCUAUAAUAGG------GGAGU .....((((((...........((((..((((((((((......))))))))))...))))..((((...))))))))))...((((.((((.(.....).)))).)))------).... ( -30.60) >DroEre_CAF1 8809 113 - 1 CUGACUGUAAUCAUAUUUUUUGGCAUAAAUGGCCACAACCACAUUGGUGGCCGUCUGGUGCCAGUCGUAGCGGCACUGCACAACCUUUUUGUCAUCAUUCUAAA-AAGG------AGAGU ..(((((....)).......((((((..((((((((..........))))))))...)))))))))((((.....))))....((((((((.........))))-))))------..... ( -32.10) >DroWil_CAF1 6997 117 - 1 CUGGGCAUAGUGAUAUUUUUUGGCAUAGAUAGUGACCACCACAUUGGUAGCGGUCUGAUGCCAAUCAUGGCGGCAUUGAACAACCUUUUUAUCCUCGUUCUGAA-AUUCGUAA--UGAGU ..((((...(((((......((((((((((.((.((((......)))).)).)))).))))))))))).))(........)..))........((((((.((..-...)).))--)))). ( -28.80) >DroYak_CAF1 8800 112 - 1 CUGACUGUAAUCAUACUUUUUGGCAUAAAUGGCCACUACCACAUUGGUGGCCGUCUGGUGCCAGUCGUAGCGGCACUGCACAACCUUUUUGUCGUCGUUCUGA--AAGU------AGAGU ..........((.(((((.(((((((..((((((((((......))))))))))...)))))))(((.((((((((..............)).)))))).)))--))))------))).. ( -36.44) >DroMoj_CAF1 6622 119 - 1 UUCCGCGUAAUGAUAUUUUUUGGCAUAGACGGCGACAACAACAUUUGUAGCUGUCUGAUGCCAAUCAUAUCGGCAUUGAAUAACCUUUUUGUCAUCAUCCUGCA-AAACAUUAAGAGAUU ....(((..(((((.....((((((((((((((.((((......)))).))))))).))))))).......((((..((......))..)))))))))..))).-............... ( -34.30) >DroAna_CAF1 6983 109 - 1 CUGAGCGUAAUGGUAUUUUUUGGCAUAAAUGGCCACCACAACGUUUGUGGCGGUCUGGUGCCAAUCGUAGCGACACUGCACAACCUUUUUGUCCUCAUUCUGAA-AAA----------GU .((((..(((.(((.....(((((((....((((.(((((.....))))).))))..)))))))..((((.....))))...)))...)))..)))).......-...----------.. ( -34.40) >consensus CUGACCGUAAUCAUAUUUUUUGGCAUAAAUGGCCACCACCACAUUGGUGGCCGUCUGGUGCCAAUCGUAGCGGCACUGCACAACCUUUUUGUCAUCAUUCUGAA_AAGC______AGAGU .((((........(((...(((((((..((((((((((......))))))))))...)))))))..)))..((((..............))))))))....................... (-22.87 = -22.37 + -0.49)



| Location | 20,012,960 – 20,013,080 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.89 |
| Mean single sequence MFE | -34.40 |
| Consensus MFE | -20.22 |
| Energy contribution | -20.95 |
| Covariance contribution | 0.73 |
| Combinations/Pair | 1.45 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.59 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.520519 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20012960 120 + 27905053 UGCAAUGUCGCUACGACUGGCACCAGACGGCCACCAAUGUGGUGAUGGCCAUUUAUGCUAAAAAAUACGAUUACAGUCAGAGCGUGAUAGAGCUGAACCCCAUCAGGCUGCACGUCAACC (((...((((...))))..)))...((((((((((.....))))..((((...(((........))).(((.....((((..(......)..)))).....))).)))))).)))).... ( -31.80) >DroPse_CAF1 8989 120 + 1 UGCAGUGCCGAUAUGAUUGGCAUCAAACGGCCACAAAUGUGGUGGUGGCCAUUUACGCCAAGAAGUAUCAUUAUGCCCAGAGUGUGAUAGAAAUAAAUCCUAUCCGUCUGCAUGUUAAGC (((((.((.((((.(((((((.....((.(((((....))))).)).)))..((((((......((((....)))).....))))))........)))).)))).)))))))........ ( -33.60) >DroGri_CAF1 10747 120 + 1 UCACAUGCCGCUACGAUUGGCAUCAGACGGCCACAAAUGUGGUUGUCGCAGUCUAUGCCAAAAAAUAUCAUUACGCCCAAAGCGUGCUGGAAGUGAAUCCGAUUCGUCUGCACAUAAUGC .........((.(((((((((((..(((((((((....))))))))).......))))))).....(((..(((((.....)))))..(((......))))))))))..))......... ( -35.40) >DroWil_CAF1 7034 120 + 1 UUCAAUGCCGCCAUGAUUGGCAUCAGACCGCUACCAAUGUGGUGGUCACUAUCUAUGCCAAAAAAUAUCACUAUGCCCAGAGUAUAGUGGAGGUGAAUCCCAUAAGGCUCCAUGUUAAUC ....(((..(((....(((((((..(((((((((....))))))))).......)))))))......(((((((((.....))))))))).((......))....)))..)))....... ( -36.90) >DroAna_CAF1 7012 120 + 1 UGCAGUGUCGCUACGAUUGGCACCAGACCGCCACAAACGUUGUGGUGGCCAUUUAUGCCAAAAAAUACCAUUACGCUCAGAGUCUGGUGGAAAUAAAUCCUAUUCGGCUGCAUGUCAAUU ((((((...((((....))))((((((((((((((.....))))))(((.......)))....................).)))))))(((......)))......))))))........ ( -35.10) >DroPer_CAF1 9215 120 + 1 UGCAGUGCCGAUAUGAUUGGCAUCAAACGGCCACAAAUGUGGUGGUGGCCAUUUACGCCAAGAAGUAUCAUUAUGCCCAGAGUGUGAUAGAAAUAAAUCCUAUCCGUCUGCAUGUUAAGC (((((.((.((((.(((((((.....((.(((((....))))).)).)))..((((((......((((....)))).....))))))........)))).)))).)))))))........ ( -33.60) >consensus UGCAAUGCCGCUACGAUUGGCAUCAGACGGCCACAAAUGUGGUGGUGGCCAUUUAUGCCAAAAAAUAUCAUUACGCCCAGAGUGUGAUAGAAAUAAAUCCUAUCCGGCUGCAUGUUAAGC ....((((.(((....(((((((.(((..(((((..........)))))..))))))))))........(((((((.....))))))).................))).))))....... (-20.22 = -20.95 + 0.73)



| Location | 20,012,960 – 20,013,080 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.89 |
| Mean single sequence MFE | -39.73 |
| Consensus MFE | -20.64 |
| Energy contribution | -20.12 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.47 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.52 |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.953828 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20012960 120 - 27905053 GGUUGACGUGCAGCCUGAUGGGGUUCAGCUCUAUCACGCUCUGACUGUAAUCGUAUUUUUUAGCAUAAAUGGCCAUCACCACAUUGGUGGCCGUCUGGUGCCAGUCGUAGCGACAUUGCA (((((.....)))))((((((((.....))))))))((((.(((((((((.........)))((((..((((((((((......))))))))))...)))))))))).))))........ ( -46.10) >DroPse_CAF1 8989 120 - 1 GCUUAACAUGCAGACGGAUAGGAUUUAUUUCUAUCACACUCUGGGCAUAAUGAUACUUCUUGGCGUAAAUGGCCACCACCACAUUUGUGGCCGUUUGAUGCCAAUCAUAUCGGCACUGCA ..........((((..(((((((.....)))))))....)))).(((...(((((....(((((((((((((((((..........)))))))))).)))))))...)))))....))). ( -40.20) >DroGri_CAF1 10747 120 - 1 GCAUUAUGUGCAGACGAAUCGGAUUCACUUCCAGCACGCUUUGGGCGUAAUGAUAUUUUUUGGCAUAGACUGCGACAACCACAUUUGUGGCCGUCUGAUGCCAAUCGUAGCGGCAUGUGA ...((((((((..((((((((((......)))...((((.....))))...))).....(((((((((((.((.((((......)))).)).)))).)))))))))))....)))))))) ( -42.20) >DroWil_CAF1 7034 120 - 1 GAUUAACAUGGAGCCUUAUGGGAUUCACCUCCACUAUACUCUGGGCAUAGUGAUAUUUUUUGGCAUAGAUAGUGACCACCACAUUGGUAGCGGUCUGAUGCCAAUCAUGGCGGCAUUGAA ............(((((((((..........((((((.(.....).)))))).......(((((((((((.((.((((......)))).)).)))).))))))))))))).)))...... ( -33.10) >DroAna_CAF1 7012 120 - 1 AAUUGACAUGCAGCCGAAUAGGAUUUAUUUCCACCAGACUCUGAGCGUAAUGGUAUUUUUUGGCAUAAAUGGCCACCACAACGUUUGUGGCGGUCUGGUGCCAAUCGUAGCGACACUGCA ........(((((.......(((......)))............((((.(((((......((((((....((((.(((((.....))))).))))..))))))))))).))).).))))) ( -36.60) >DroPer_CAF1 9215 120 - 1 GCUUAACAUGCAGACGGAUAGGAUUUAUUUCUAUCACACUCUGGGCAUAAUGAUACUUCUUGGCGUAAAUGGCCACCACCACAUUUGUGGCCGUUUGAUGCCAAUCAUAUCGGCACUGCA ..........((((..(((((((.....)))))))....)))).(((...(((((....(((((((((((((((((..........)))))))))).)))))))...)))))....))). ( -40.20) >consensus GCUUAACAUGCAGACGGAUAGGAUUCAUUUCCAUCACACUCUGGGCAUAAUGAUAUUUUUUGGCAUAAAUGGCCACCACCACAUUUGUGGCCGUCUGAUGCCAAUCAUAGCGGCACUGCA ........(((((.......(((......)))............((...((((......(((((((((((.(((((..........))))).)))).)))))))))))....)).))))) (-20.64 = -20.12 + -0.52)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:47:21 2006