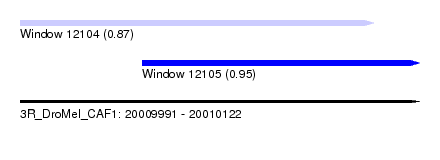
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 20,009,991 – 20,010,122 |
| Length | 131 |
| Max. P | 0.954330 |

| Location | 20,009,991 – 20,010,107 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 74.93 |
| Mean single sequence MFE | -16.97 |
| Consensus MFE | -9.88 |
| Energy contribution | -9.77 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.868679 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
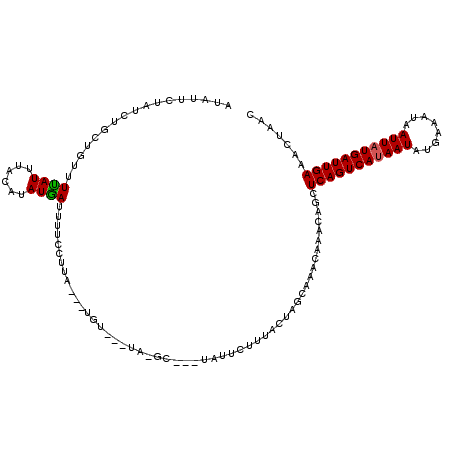
>3R_DroMel_CAF1 20009991 116 + 27905053 AUAUUCUAUCUGCUGUUUUAUUUAUAUAUAAUUUUCCUUACAAUGUGCAUAAGC---UAUUCUUUACUAGGAAACAAGCAGUUCAGUCACAAUAUGAAAUAAUUAUGAUUGAAACUAAC .........(((((...((((......)))).((((((...((((.((....))---)))).......))))))..)))))((((((((.(((........))).))))))))...... ( -18.10) >DroSec_CAF1 4004 101 + 1 AUAUUCUAUCUGCUGUUUUAUUUACAUAUGAUUUUCCUUA---UGU---------------CUUUACUAGCAAACAAACAGCUCAGUCAUAAUAUGAAAUAAUUAUGAUUGAAACUAAC ...........(((((((.....(((((.(......).))---)))---------------((.....)).....)))))))(((((((((((........)))))))))))....... ( -18.10) >DroSim_CAF1 3962 100 + 1 ----------------AUCAUUUACAUAUGAUUUUCCUUA---UGUAUUUAUGCACUUAUUCUUUACUAGCAAACAAACAGCUCAGUCAUAAUAUGAAAUAAUUAUGAUUGAAACUAAC ----------------..(((.((((((.(......).))---))))...))).............................(((((((((((........)))))))))))....... ( -14.70) >consensus AUAUUCUAUCUGCUGUUUUAUUUACAUAUGAUUUUCCUUA___UGU___UA_GC___UAUUCUUUACUAGCAAACAAACAGCUCAGUCAUAAUAUGAAAUAAUUAUGAUUGAAACUAAC .................((((......))))...................................................(((((((((((........)))))))))))....... ( -9.88 = -9.77 + -0.11)


| Location | 20,010,031 – 20,010,122 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 66.00 |
| Mean single sequence MFE | -11.44 |
| Consensus MFE | -6.28 |
| Energy contribution | -7.52 |
| Covariance contribution | 1.24 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.55 |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.954330 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
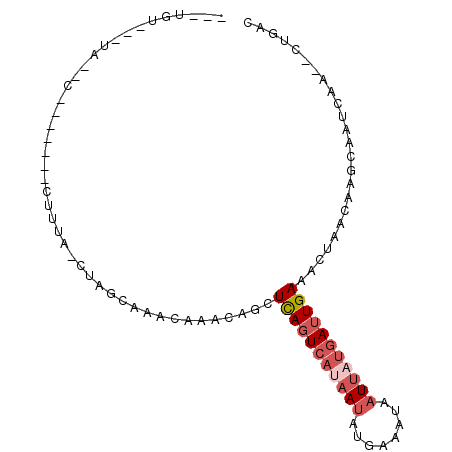
>3R_DroMel_CAF1 20010031 91 + 27905053 CAAUGUGCAUAAGC---UAUUCUUUA-CUAGGAAACAAGCAGUUCAGUCACAAUAUGAAAUAAUUAUGAUUGAAACUAACAAGCAAUCAA--CUGAC .....(((....((---(.(((((..-..)))))...)))((((((((((.(((........))).)))))).)))).....))).....--..... ( -15.90) >DroSec_CAF1 4044 76 + 1 ---UGU---------------CUUUA-CUAGCAAACAAACAGCUCAGUCAUAAUAUGAAAUAAUUAUGAUUGAAACUAACAAGCAAUCAA--CUGAC ---.((---------------(....-...((.........))(((((((((((........))))))))))).................--..))) ( -11.80) >DroSim_CAF1 3986 91 + 1 ---UGUAUUUAUGCACUUAUUCUUUA-CUAGCAAACAAACAGCUCAGUCAUAAUAUGAAAUAAUUAUGAUUGAAACUAACAAGCAAUCAA--CUGAC ---........(((............-...((.........))(((((((((((........))))))))))).........))).....--..... ( -14.00) >DroEre_CAF1 5893 85 + 1 GCAUUUAUUUAUCC--------UACA-CUAACCACCAAGC-UCUCAGUCAUAAUAUUAAAUAAUUAUGAUUGAAACUAACAAGCAAUCAA--CUGAC ((............--------....-((........)).-..(((((((((((........))))))))))).........))......--..... ( -11.00) >DroAna_CAF1 4393 74 + 1 ----------------------UACAAAUUGCCAAAAAUU-ACUUAGUACAAACACUAAAAAAUCAUAAAUGAAAAUUCCCAAGAGUUAGCGCCGAC ----------------------........((........-..(((((......)))))...............(((((....))))).))...... ( -4.50) >consensus ___UGU___UA__C_______CUUUA_CUAGCAAACAAACAGCUCAGUCAUAAUAUGAAAUAAUUAUGAUUGAAACUAACAAGCAAUCAA__CUGAC ...........................................(((((((((((........)))))))))))........................ ( -6.28 = -7.52 + 1.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:47:18 2006