
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 19,985,641 – 19,985,755 |
| Length | 114 |
| Max. P | 0.609952 |

| Location | 19,985,641 – 19,985,755 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.89 |
| Mean single sequence MFE | -36.32 |
| Consensus MFE | -20.09 |
| Energy contribution | -21.74 |
| Covariance contribution | 1.64 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.609952 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
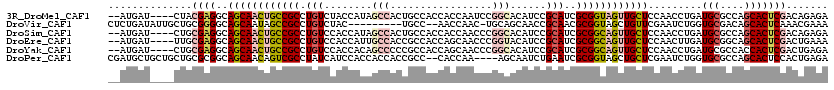
>3R_DroMel_CAF1 19985641 114 - 27905053 --AUGAU----CUACGAGGCAGCAACUGCCGCCUGUCUACCAUAGCCACUGCCACCACCAAUCCGGCACAUCCGCAUCGCGGUAGUUGCUCCAACCUGAUGCGCCAGCACUCGACAGAGA --....(----((.(((((.((((((((((((((((.....))))....((((...........))))..........)))))))))))).).......(((....)))))))..))).. ( -34.60) >DroVir_CAF1 3901 108 - 1 CUCUGAUAUUGCUGCGGGGCAGCAAUAGCCGCCUGUCUAC---------UGCC--AACCAAC-UGCAGCAACCGCAACGCGGUAGCUGUUCGAAUCUGGUGCGACAGCACUCAAACGAAA ...((((((((((((...)))))))))...((.((((...---------((((--(......-.(((((.(((((...))))).))))).......))))).))))))..)))....... ( -37.44) >DroSim_CAF1 3869 114 - 1 --AUGAU----CUGCGAGGCAGCAACUGCCGCCUGUCCACCAUAGCCACUGCCACCACCAACCCGGCACAUCCGCAUCGCGGCAGUUGCUCCAACCUGAUGCGCCAGCACUCGACAGAGA --....(----((((((((.((((((((((((((((.....))))....((((...........))))..........)))))))))))).).......(((....))))))).)))).. ( -41.30) >DroEre_CAF1 3922 114 - 1 --AUGAU----UUGCGAGGCAGCAACUGCCGCCUGUCCACCAUUGCCACCGCCACCAGCAACCCGGUACAUCCGCAUCGCGGCAGUUGCUCCAACUUGAUGCGGCAGCACUCGACUGAAA --.....----...(((((.((((((((((((..........((((...........))))..(((.....)))....)))))))))))).).......(((....)))))))....... ( -35.70) >DroYak_CAF1 3868 114 - 1 --AUGAU----CUGCGAGGCAGCAACUGCCGCCUGUCCACCACAGCCCCCGCCACCAGCAACCCGGCACAUCCGCAUCGCGGCAGUUGCUCCAACCUGAUGCGCCACCACUCGACUGAGA --...((----(.....((.((((((((((((((((.....)))).....(((...........)))...........)))))))))))))).....))).........(((....))). ( -34.90) >DroPer_CAF1 9224 114 - 1 CGAUGCUGCUGCUGCGCGGCAGCAACAGUCGCCUAUCAUCCACCACCACCGCC--CACCAA----AGCAAUCUGAAUCGCGGUAGCUGCUCGAAUCUGGUGCGCCAGCACUCCACUGAGA (((.((.(((((((((((((.......)))))..................((.--......----.))..........)))))))).))))).....(((((....)))))......... ( -34.00) >consensus __AUGAU____CUGCGAGGCAGCAACUGCCGCCUGUCCACCAUAGCCACCGCCACCACCAACCCGGCACAUCCGCAUCGCGGCAGUUGCUCCAACCUGAUGCGCCAGCACUCGACUGAGA ..............((((..((((((((((((.(((........(((.................)))......)))..)))))))))))).........(((....)))))))....... (-20.09 = -21.74 + 1.64)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:47:07 2006