| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 19,953,097 – 19,953,195 |
| Length | 98 |
| Max. P | 0.595437 |

| Location | 19,953,097 – 19,953,195 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 82.49 |
| Mean single sequence MFE | -29.88 |
| Consensus MFE | -21.53 |
| Energy contribution | -22.06 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.595437 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19953097 98 + 27905053 A---UAAGAUCUAUUUCAACUCCC-----UCACU-UUACAGAUCUCAAUGCUGCGCGAGCAGUUGGCCCAGGAGGCCAAACGGCGACAGCAGUACAUUCUACGCAGC .---..((((((............-----.....-....))))))....(((((((..((..((((((.....))))))...))....)).(((.....)))))))) ( -27.00) >DroVir_CAF1 16719 95 + 1 C---ACAGAACU---GCAGC-----UGAGCUCCUC-CACAGAUCUCCAUGCUGCGCGAGCAGCUGGCUCAGGAGGCCAAGCGGCGACAGCAAUAUAUACUGCGCAGC .---......((---((.((-----((....((((-(...((...(((.(((((....)))))))).)).))))).....))))....(((........))))))). ( -36.40) >DroSec_CAF1 12781 98 + 1 A---UAAGAUCUACAUCAAGUUCC-----UUACU-UUUCAGAUCUCUAUGCUGCGCGAGCAGUUGGCCCAGGAGGCCAAACGGCGACAGCAGUACAUUCUACGCAGC .---..((((((.....((((...-----..)))-)...))))))....(((((((..((..((((((.....))))))...))....)).(((.....)))))))) ( -29.70) >DroSim_CAF1 12515 98 + 1 A---UAAGAUCUACAUAAAGUUCC-----UUACU-UUUCAGAUCUCUAUGCUGCGCGAGCAGUUGGCCCAGGAGGCCAAACGGCGACAGCAGUACAUUCUACGCAGC .---..((((((....(((((...-----..)))-))..))))))....(((((((..((..((((((.....))))))...))....)).(((.....)))))))) ( -31.60) >DroEre_CAF1 12835 98 + 1 A---UAAUAUCUACAUGCAGUACC-----CCACU-UUUCAGAUCUCUAUGCUGCGCGAGCAGCUGGCUCAGGAGGCCAAACGGCGGCAGCAGUACAUUCUACGCAGC .---...........(((.((.((-----((...-..............(((((....)))))(((((.....)))))...)).))..)).(((.....)))))).. ( -25.70) >DroYak_CAF1 12700 103 + 1 AAAGUAAUAUCUACAUCAUGUACCCU---CCACU-UUUCAGAUCUCUAUGCUGCGCGAGCAGUUGGCUCAGGAGGCCAAACGGCGGCAGCAGUACAUUCUGCGCAGC (((((......(((.....)))....---..)))-))............(((((((..((..((((((.....))))))...)).)).((((......))))))))) ( -28.90) >consensus A___UAAGAUCUACAUCAAGUACC_____CCACU_UUUCAGAUCUCUAUGCUGCGCGAGCAGUUGGCCCAGGAGGCCAAACGGCGACAGCAGUACAUUCUACGCAGC ......((((((...........................))))))....(((((((..((..((((((.....))))))...))....)).(((.....)))))))) (-21.53 = -22.06 + 0.53)


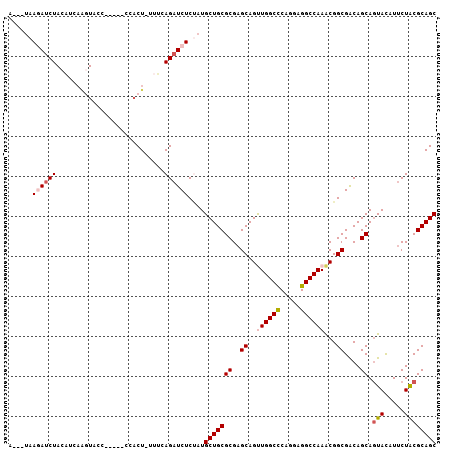
| Location | 19,953,097 – 19,953,195 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 82.49 |
| Mean single sequence MFE | -33.97 |
| Consensus MFE | -25.92 |
| Energy contribution | -25.81 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.593517 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19953097 98 - 27905053 GCUGCGUAGAAUGUACUGCUGUCGCCGUUUGGCCUCCUGGGCCAACUGCUCGCGCAGCAUUGAGAUCUGUAA-AGUGA-----GGGAGUUGAAAUAGAUCUUA---U (((((((((......))))....((.(((.((((.....))))))).))....)))))..((((((((((..-.....-----..........))))))))))---. ( -33.53) >DroVir_CAF1 16719 95 - 1 GCUGCGCAGUAUAUAUUGCUGUCGCCGCUUGGCCUCCUGAGCCAGCUGCUCGCGCAGCAUGGAGAUCUGUG-GAGGAGCUCA-----GCUGC---AGUUCUGU---G (((((((((((.....))))))....(((.(((((((.((.((((((((....))))).)))...))...)-)))..))).)-----)).))---))).....---. ( -38.60) >DroSec_CAF1 12781 98 - 1 GCUGCGUAGAAUGUACUGCUGUCGCCGUUUGGCCUCCUGGGCCAACUGCUCGCGCAGCAUAGAGAUCUGAAA-AGUAA-----GGAACUUGAUGUAGAUCUUA---U (((((((((......))))....((.(((.((((.....))))))).))....)))))...((((((((..(-(((..-----...))))....)))))))).---. ( -35.80) >DroSim_CAF1 12515 98 - 1 GCUGCGUAGAAUGUACUGCUGUCGCCGUUUGGCCUCCUGGGCCAACUGCUCGCGCAGCAUAGAGAUCUGAAA-AGUAA-----GGAACUUUAUGUAGAUCUUA---U (((((((((......))))....((.(((.((((.....))))))).))....)))))...((((((((.((-(((..-----...)))))...)))))))).---. ( -35.90) >DroEre_CAF1 12835 98 - 1 GCUGCGUAGAAUGUACUGCUGCCGCCGUUUGGCCUCCUGAGCCAGCUGCUCGCGCAGCAUAGAGAUCUGAAA-AGUGG-----GGUACUGCAUGUAGAUAUUA---U .((((((.(...((((((((((((((....))).....((((.....))))).))))).(((....)))...-.....-----)))))..)))))))......---. ( -29.20) >DroYak_CAF1 12700 103 - 1 GCUGCGCAGAAUGUACUGCUGCCGCCGUUUGGCCUCCUGAGCCAACUGCUCGCGCAGCAUAGAGAUCUGAAA-AGUGG---AGGGUACAUGAUGUAGAUAUUACUUU (((((((((......)))).((.((.(((.((((....).)))))).))..)))))))......(((((...-.(((.---......)))....)))))........ ( -30.80) >consensus GCUGCGUAGAAUGUACUGCUGUCGCCGUUUGGCCUCCUGAGCCAACUGCUCGCGCAGCAUAGAGAUCUGAAA_AGUAA_____GGAACUUGAUGUAGAUCUUA___U (((((((((......)))).((.((.(((.(((.......)))))).))..)))))))...((((((((.........................))))))))..... (-25.92 = -25.81 + -0.11)

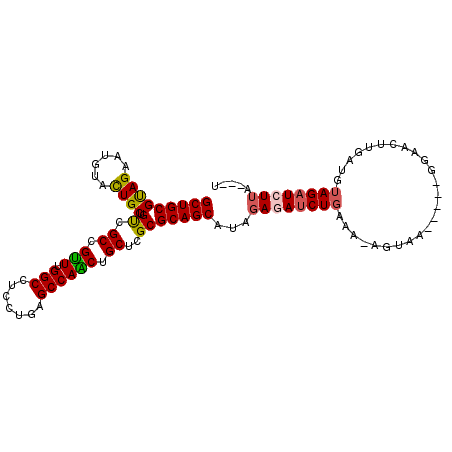

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:46:57 2006