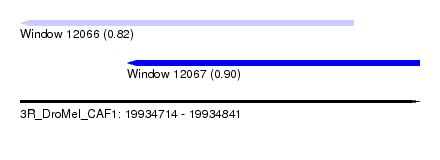
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 19,934,714 – 19,934,841 |
| Length | 127 |
| Max. P | 0.900429 |

| Location | 19,934,714 – 19,934,820 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 78.41 |
| Mean single sequence MFE | -24.11 |
| Consensus MFE | -17.62 |
| Energy contribution | -18.46 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.819177 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
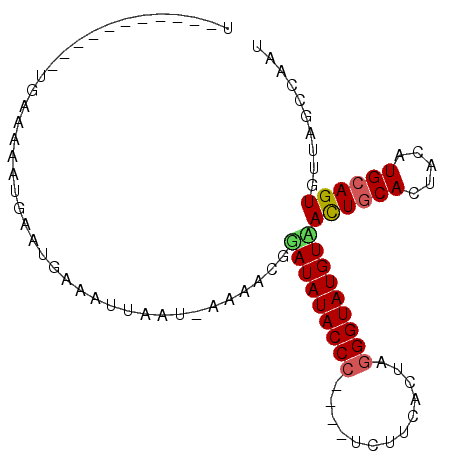
>3R_DroMel_CAF1 19934714 106 - 27905053 AAUGAAAUUAAU-G--AAACAUAUAUACCC----ACUUCAGUAGGGUAUGUAACUGCACUACGUGCAGUGCUGGCCAAUGUCAAAAUUCUUCUUCGACCUACAGCACUGGUCG ............-.--.....(((((((((----((....)).)))))))))(((((((...)))))))...(((((.(((......((......))......))).))))). ( -29.30) >DroSec_CAF1 47220 107 - 1 AAUGAAAUUGAUUG--AAACGGAUAUACCC----UCUUCACUAGGGUAUGUUACUGCACUACAUGCAGUGUUAGCCAAUGUCAAAAUUCAUCUUCGACCUACAGCACUGGACG ..((((..((((((--(...((((((((((----(.......)))))))))(((((((.....)))))))....))....))))...)))..)))).((.........))... ( -25.20) >DroSim_CAF1 48470 106 - 1 AAUGAAAUAAAU-A--UAAUGAAUAUACCC----UCUUCACUAGGGUAUGUUAUUGCACUACAUGCAGUGUUAGCCAAUGUCAAAAUUCCUCUUCGACCUACAGCACUGGUCG ..(((.......-.--((((.(((((((((----(.......))))))))))((((((.....)))))))))).......)))...........(((((.........))))) ( -23.66) >DroEre_CAF1 48470 101 - 1 A---------AU-A--AAAUGGAUAUACCCCCUUCCUUCCCUGUGGUAUGUAACUGCACUACGUGAAGUGUUGGCCAAUGCCAAAAUUCCUCUUCGACCUACAGCACUGGUCG .---------..-.--....(((........((((......((..((.....))..))......))))..(((((....)))))...)))....(((((.........))))) ( -20.50) >DroYak_CAF1 49408 99 - 1 A---------AU-AAUAAACUGAUAUACCU----UCUGUACUACGGUAUGUGACUGCACAAUUUGCAGUGUUGGCUAAUGCCGAAAUUCCUCAUCGACCUACAACACUAGUCG .---------..-.........(((((((.----..........)))))))(((((((.....)))(((((((((....)))((......))..........)))))))))). ( -21.90) >consensus AAUGAAAU__AU_A__AAACGGAUAUACCC____UCUUCACUAGGGUAUGUAACUGCACUACAUGCAGUGUUGGCCAAUGUCAAAAUUCCUCUUCGACCUACAGCACUGGUCG .....................((((((((...............))))))))((((((.....)))))).(((((....)))))..........(((((.........))))) (-17.62 = -18.46 + 0.84)

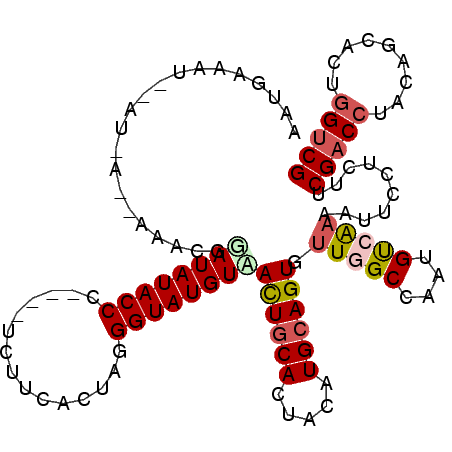
| Location | 19,934,748 – 19,934,841 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 70.64 |
| Mean single sequence MFE | -18.32 |
| Consensus MFE | -12.55 |
| Energy contribution | -12.79 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.900429 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19934748 93 - 27905053 UUUAACUCGAGUUGAAAAAUUAAUGAAAUUAAU-GAAACAUAUAUACCC----ACUUCAGUAGGGUAUGUAACUGCACUACGUGCAGUGCUGGCCAAU ((((((....)))))).................-......(((((((((----((....)).)))))))))(((((((...))))))).......... ( -23.70) >DroSec_CAF1 47254 83 - 1 U-----------UGAAAAAUGAAUGAAAUUGAUUGAAACGGAUAUACCC----UCUUCACUAGGGUAUGUUACUGCACUACAUGCAGUGUUAGCCAAU .-----------...............((((..(((....(((((((((----(.......))))))))))((((((.....)))))).)))..)))) ( -20.20) >DroSim_CAF1 48504 82 - 1 A-----------UGAAAAAUGAAUGAAAUAAAU-AUAAUGAAUAUACCC----UCUUCACUAGGGUAUGUUAUUGCACUACAUGCAGUGUUAGCCAAU .-----------.......((..(((.......-......(((((((((----(.......))))))))))((((((.....)))))).)))..)).. ( -16.10) >DroEre_CAF1 48504 88 - 1 UUCAGCUUGAGUCUUAAAAUGA---------AU-AAAAUGGAUAUACCCCCUUCCUUCCCUGUGGUAUGUAACUGCACUACGUGAAGUGUUGGCCAAU ((((..((((...))))..)))---------).-....((((((((((...............)))))))..(.(((((......))))).).))).. ( -13.26) >consensus U___________UGAAAAAUGAAUGAAAUUAAU_AAAACGGAUAUACCC____UCUUCACUAGGGUAUGUAACUGCACUACAUGCAGUGUUAGCCAAU ........................................(((((((((.............)))))))))((((((.....)))))).......... (-12.55 = -12.79 + 0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:46:41 2006