
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 19,904,212 – 19,904,359 |
| Length | 147 |
| Max. P | 0.790847 |

| Location | 19,904,212 – 19,904,322 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 79.73 |
| Mean single sequence MFE | -37.86 |
| Consensus MFE | -24.57 |
| Energy contribution | -24.90 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.639674 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19904212 110 + 27905053 -AUAGCUGACCUCUGCGCU---GCCCCAAACC-CUUU--GCGCUGGCAAAUUUCAGUCCAUCACAUUUUGCAUGCAACCCGCCGAUCGGGGCUAAUGCAAUCAAGUGGGCGUGGUCA -......((((.(.((((.---..........-....--))))............((((((......((((((....((((.....))))....))))))....))))))).)))). ( -33.46) >DroSec_CAF1 17637 111 + 1 -AUGGCUGACCUCUGCGC----GCUCCAAAAC-CUUUUGGGGGUGGCAAAUUUCAGCCCAUCGCAUUUUGCAUGCAACCCGUCGAUCGGGGCUAAUGCAAUCAAGUGGGCGUGGUCA -......((((.((((((----.(((((((..-..))))))))).))).......((((((......((((((....((((.....))))....))))))....))))))).)))). ( -42.70) >DroSim_CAF1 18515 111 + 1 -AUGGCUGACCUCUGCGC----GCUCCAAACC-CUUUUGGGGGUGGCAAAUUUCAGCCCAUCGCAUUUUGCAUGCAACCCGCCGAUCGGGGCUAAUGCAAUCAAGUGGGCGUGGUCA -......((((.((((.(----(((((.....-......))))))))).......((((((......((((((....((((.....))))....))))))....))))))).)))). ( -41.70) >DroEre_CAF1 17965 106 + 1 -AUGGCUGACCUCUGCCCCAU---------GCCCCUU-UGGGGUGGCAAAUUUCAGCCCAUCGCAUUUUGCAUGCAACCAGCCGAUCGGGGCUAAUGCAAUCAAGUGGGCGUGGUCA -......((((.(((((....---------(((((..-.))))))))).......((((((.((.....)).((((...((((......))))..)))).....))))))).)))). ( -40.40) >DroYak_CAF1 18389 116 + 1 -AUGGCUGACCUCUGCCCCAAAGCUCCAAAGCUCCAGAGGAGGUGGCAAAUUUCCGUCCAUCGCAUUUUGCAUGCUACCAGCCGAUCGGGGCUAAUGCAAUCAAGUGGGCGUGGUCA -..((((..((((((......((((....)))).)))))).(((((((.......(.....)((.....)).)))))))))))(((((.(.(((.((....))..))).).))))). ( -37.10) >DroAna_CAF1 17952 106 + 1 CACAACCCACCUCUCCUA----GCCCCG---CCUAGU-UCGCCUGCCAAC-CUC--UUCACGGGCUUUUGCAUGCAACCCGCCUAUUGGGGCUAAUGCAAUCAAGUGGGCGUGGUCA ..................----(((.((---((((.(-(.(((((.....-...--....)))))..((((((....((((.....))))....))))))..)).)))))).))).. ( -31.80) >consensus _AUGGCUGACCUCUGCGC____GCUCCAAACC_CUUU_GGGGGUGGCAAAUUUCAGCCCAUCGCAUUUUGCAUGCAACCCGCCGAUCGGGGCUAAUGCAAUCAAGUGGGCGUGGUCA .......((((.((((.............................))).......((((((......((((((....((((.....))))....))))))....))))))).)))). (-24.57 = -24.90 + 0.33)



| Location | 19,904,245 – 19,904,359 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 77.80 |
| Mean single sequence MFE | -30.48 |
| Consensus MFE | -22.45 |
| Energy contribution | -22.78 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.790847 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19904245 114 + 27905053 CGCUGGCAAAUUUCAGUCCAUCACAUUUUGCAUGCAACCCGCCGAUCGGGGCUAAUGCAAUCAAGUGGGCGUGGUCAGCAAAUAGCUGGUAUGAACAAGAAGAAAAAAAUAACA--- .((((((........((((((......((((((....((((.....))))....))))))....))))))...))))))...................................--- ( -30.00) >DroSec_CAF1 17671 103 + 1 GGGUGGCAAAUUUCAGCCCAUCGCAUUUUGCAUGCAACCCGUCGAUCGGGGCUAAUGCAAUCAAGUGGGCGUGGUCAGCAAAUAGCUGGUAC----------AAA-AAAUAACA--- ..(((.......(((((((((......((((((....((((.....))))....))))))....)))))).)))(((((.....))))))))----------...-........--- ( -31.30) >DroSim_CAF1 18549 104 + 1 GGGUGGCAAAUUUCAGCCCAUCGCAUUUUGCAUGCAACCCGCCGAUCGGGGCUAAUGCAAUCAAGUGGGCGUGGUCAGCAAAUAGCUGGUGC----------AAAAAAAUAACA--- .((((((........).)))))...((((((((...(((((((.((..(.((....))...)..)).)))).)))((((.....))))))))----------))))........--- ( -32.70) >DroEre_CAF1 17994 114 + 1 GGGUGGCAAAUUUCAGCCCAUCGCAUUUUGCAUGCAACCAGCCGAUCGGGGCUAAUGCAAUCAAGUGGGCGUGGUCAGCAAAUAGCUGGUACGAGCAACAACAAAAAAAUAAAA--- ..((.((.....(((((((((.((.....)).((((...((((......))))..)))).....)))))).)))(((((.....))))).....)).))...............--- ( -30.90) >DroYak_CAF1 18428 101 + 1 AGGUGGCAAAUUUCCGUCCAUCGCAUUUUGCAUGCUACCAGCCGAUCGGGGCUAAUGCAAUCAAGUGGGCGUGGUCAGCAAAUAGCUGCUAUGAAGAACAA---------------- .(((((((.......(.....)((.....)).)))))))((((......))))...........((...(((((.((((.....)))))))))....))..---------------- ( -28.90) >DroAna_CAF1 17984 96 + 1 GCCUGCCAAC-CUC--UUCACGGGCUUUUGCAUGCAACCCGCCUAUUGGGGCUAAUGCAAUCAAGUGGGCGUGGUCAGCCCA-----G--GC----------AAA-CAACAACAGAG (((((.....-...--....))))).(((((..((.(((((((((((...((....)).....)))))))).)))..))...-----.--))----------)))-........... ( -29.10) >consensus GGGUGGCAAAUUUCAGCCCAUCGCAUUUUGCAUGCAACCCGCCGAUCGGGGCUAAUGCAAUCAAGUGGGCGUGGUCAGCAAAUAGCUGGUAC__________AAA_AAAUAACA___ ...............((((((......((((((....((((.....))))....))))))....)))))).....((((.....))))............................. (-22.45 = -22.78 + 0.33)

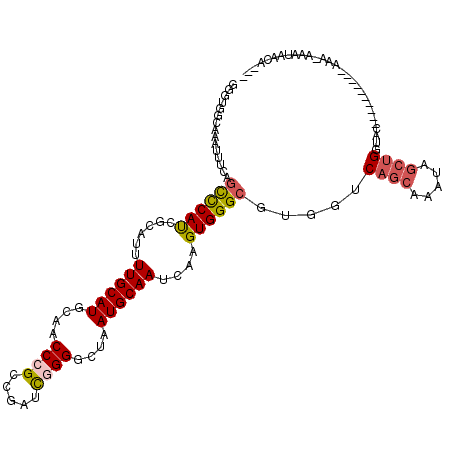

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:46:34 2006