| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 19,887,375 – 19,887,482 |
| Length | 107 |
| Max. P | 0.519952 |

| Location | 19,887,375 – 19,887,482 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 80.61 |
| Mean single sequence MFE | -28.40 |
| Consensus MFE | -19.48 |
| Energy contribution | -19.40 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.69 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.519952 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
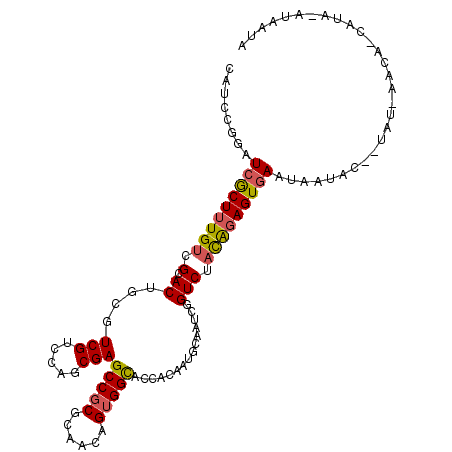
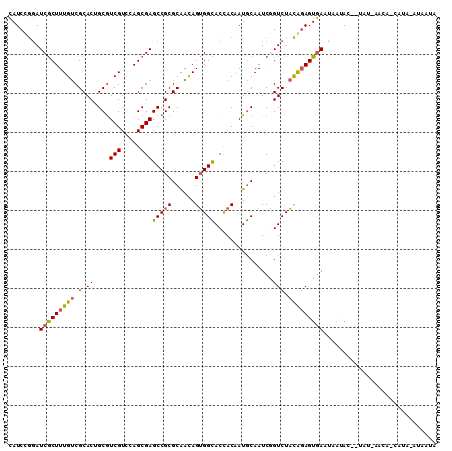
>3R_DroMel_CAF1 19887375 107 - 27905053 CAUCCGGAUCGCUUUGUCGCACUGCGUCGUCCAGCGAGCCGCGCAACAGUGGUACCACGAUGCAAUCGGUCUACAGAGUGAAUAAUACAUUAU-AAGA-CAGA-GUAAUA .......((..(((((((....((((((((...(.(.(((((......))))).)))))))))).(((.((....)).)))............-..))-))))-)..)). ( -31.40) >DroVir_CAF1 3270 101 - 1 UAUCCACAUCACUUUGCCGCACUGCAUCGUCCAGCGAGCCGCGCAGUAGGGGUACCACAAUACAAUCCGUCUUUGGAGUAAGAAACAU--UG-UUA----AUA-CAAAU- ...........((((((.((.(.((.(((.....))))).).)).))))))(((..(((((....((((....))))((.....))))--))-)..----.))-)....- ( -20.50) >DroSec_CAF1 3254 106 - 1 CAUCUGGAUCGCUUUGUCGCACAGCGUCGUCCAGCGAGCCGCGCAACAGUGGCACCACAAUGCAAUCAGUCUACAUAGUGAAUAAUAU--UAU-AACAUUAUA-AUAAUA ...((((((((((.((...)).))))..))))))...(((((......)))))..(((.(((...........))).))).....(((--(((-(....))))-)))... ( -25.00) >DroSim_CAF1 3249 99 - 1 CAUCUGGAUCGCUUUGUCGCACAGCGUCGUCCAGCGAGCCGCGCGACAGUGGCACCACAAUGCAAUCGGUCUACAGAGUGAAUAAUAC--UA--------AUA-AUAAUA ........((((((((((((.(.((.(((.....))))).).))))).((((.(((...........)))))))))))))).......--..--------...-...... ( -28.20) >DroEre_CAF1 3262 104 - 1 CAUCCGGAUCGCUUUGUCGCACUGCGUCGUCCAGCGAGCCGCGCAACAGUGGCACUACGAUGCAAUCGGUCUACGGAGUGAAUAACAC---AU-AACG-CAAAAAUAAU- ..((((((((((......))..((((((((..((...(((((......))))).))))))))))...))))..))))(((.....)))---..-....-..........- ( -30.40) >DroYak_CAF1 3248 104 - 1 CAUCCGGAUCGCUUUGUCGCACCGCGUCGUCCAGCGAGCCGCGCAACAGUGGCACCACGAUGCAAUCGGUCUACGGAGUGAUCACAAU---GU-AACA-CAAA-AUAAUG (((...(((((((((((.(.((((((((((...(.(.(((((......))))).)))))))))....)))).)))))))))))...))---).-....-....-...... ( -34.90) >consensus CAUCCGGAUCGCUUUGUCGCACUGCGUCGUCCAGCGAGCCGCGCAACAGUGGCACCACAAUGCAAUCGGUCUACAGAGUGAAUAAUAC__UAU_AACA_CAUA_AUAAUA ........(((((((((.(.((....(((.....)))(((((......)))))...............))).)))))))))............................. (-19.48 = -19.40 + -0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:46:31 2006