
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 19,869,499 – 19,869,688 |
| Length | 189 |
| Max. P | 0.940702 |

| Location | 19,869,499 – 19,869,619 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -45.67 |
| Consensus MFE | -43.11 |
| Energy contribution | -43.00 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.641315 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19869499 120 + 27905053 CAACGGCAUGGCCAGGAGCGGCUCCGACUUUGCUACUACUCGGUAUUGUACGUAACCUAAACGUGCGUAAUGCGCUUGCAACUGCUGCAGGUGCGGAAGCUGAUGCAGCAGAGGUGGUUG ((((.((..((((......))))....(((((((.(...(((((...((((((.......))))))....((((((((((.....))))))))))...))))).).))))))))).)))) ( -45.50) >DroSec_CAF1 5278 120 + 1 CAACGGCAUGGCCAGGAGCGGCUCCGACGUUGUUACUACUCGGUAUUGUACGUAACCUAAACGUGCGUAAUGCGCUUGCAACUGCUGCAGGUGCGGAAGCUGAUGCAGCAGAGGUGGUUG (((((....((((......))))....)))))..((((((..(((((((((((.......))))))..)))))........((((((((...((....))...)))))))).)))))).. ( -45.70) >DroSim_CAF1 5280 118 + 1 CAACGGCAUGGCCAGGAGCGGCUCCGACUUUGUUACUACUCGGUA-UGUACGUAAC-UAAACGUGCGUAAUGCGCUUGCAACUGCUGCAGGUGCAGAAGCUGAUGCAGCAGAGGUGGUUG ((((.((..((((......))))....(((((((.(...((((((-(((((((...-...))))))))..((((((((((.....))))))))))...))))).).))))))))).)))) ( -45.80) >consensus CAACGGCAUGGCCAGGAGCGGCUCCGACUUUGUUACUACUCGGUAUUGUACGUAACCUAAACGUGCGUAAUGCGCUUGCAACUGCUGCAGGUGCGGAAGCUGAUGCAGCAGAGGUGGUUG ((((.((..((((......))))....(((((((.(...(((((...((((((.......))))))....((((((((((.....))))))))))...))))).).))))))))).)))) (-43.11 = -43.00 + -0.11)



| Location | 19,869,539 – 19,869,659 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.78 |
| Mean single sequence MFE | -49.53 |
| Consensus MFE | -47.82 |
| Energy contribution | -47.60 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.827849 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19869539 120 + 27905053 CGGUAUUGUACGUAACCUAAACGUGCGUAAUGCGCUUGCAACUGCUGCAGGUGCGGAAGCUGAUGCAGCAGAGGUGGUUGUGGCUGCGGCUGGGGGCGACUUCCCGGAUGCUGCAAACGG (((((((((((((.......))))))..)))))..(..(..((((((((...((....))...))))))))..)..)(((..((.....(((((((....)))))))..))..))).)). ( -48.90) >DroSec_CAF1 5318 120 + 1 CGGUAUUGUACGUAACCUAAACGUGCGUAAUGCGCUUGCAACUGCUGCAGGUGCGGAAGCUGAUGCAGCAGAGGUGGUUGUGGCUGCGGCUGGGGUCGACUUCCCGGAUGCUGCAAACGG (((((((((((((.......))))))..)))))..(..(..((((((((...((....))...))))))))..)..)(((..((.....((((((......))))))..))..))).)). ( -49.50) >DroSim_CAF1 5320 118 + 1 CGGUA-UGUACGUAAC-UAAACGUGCGUAAUGCGCUUGCAACUGCUGCAGGUGCAGAAGCUGAUGCAGCAGAGGUGGUUGUGGCUGCGGCUGGGGGCGACUUCCCGGAUGCUGCAAACGG (((((-(((((((...-...))))))))..((((((((((.....))))))))))...)))).(((((((...(..((....))..)..(((((((....))))))).)))))))..... ( -50.20) >consensus CGGUAUUGUACGUAACCUAAACGUGCGUAAUGCGCUUGCAACUGCUGCAGGUGCGGAAGCUGAUGCAGCAGAGGUGGUUGUGGCUGCGGCUGGGGGCGACUUCCCGGAUGCUGCAAACGG ((((...((((((.......))))))....((((((((((.....))))))))))...)))).(((((((...(..((....))..)..((((((......)))))).)))))))..... (-47.82 = -47.60 + -0.22)

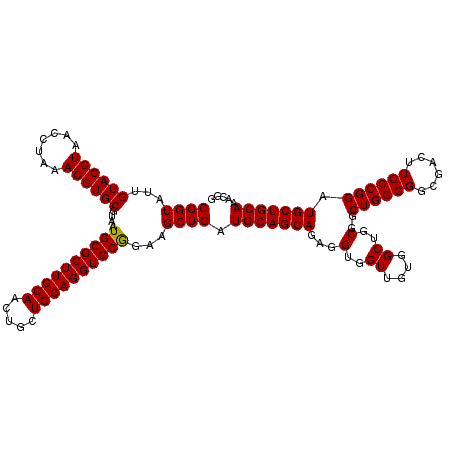

| Location | 19,869,579 – 19,869,688 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.78 |
| Mean single sequence MFE | -53.07 |
| Consensus MFE | -52.13 |
| Energy contribution | -52.13 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.940702 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19869579 109 + 27905053 ACUGCUGCAGGUGCGGAAGCUGAUGCAGCAGAGGUGGUUGUGGCUGCGGCUGGGGGCGACUUCCCGGAUGCUGCAAACGGCGUCGAAGCUGGCGUCACAUCUCAGCUCA----------- ..(((((((...((....))...)))))))(((.(((.((((((..((((((((((....))))).(((((((....)))))))..)))))..)))))).).)).))).----------- ( -53.50) >DroSec_CAF1 5358 120 + 1 ACUGCUGCAGGUGCGGAAGCUGAUGCAGCAGAGGUGGUUGUGGCUGCGGCUGGGGUCGACUUCCCGGAUGCUGCAAACGGCGUCGAAGCUGGCGUCACAUCUCAGCUCAUUGGGUCUGGU .((((((((...((....))...))))))))((((((((...((.(((.((((((......)))))).))).)).)))(((((((....))))))).)))))(((((.....)).))).. ( -55.00) >DroSim_CAF1 5358 120 + 1 ACUGCUGCAGGUGCAGAAGCUGAUGCAGCAGAGGUGGUUGUGGCUGCGGCUGGGGGCGACUUCCCGGAUGCUGCAAACGGCGUCGAAGCUGGCGUCACAUCUCAGCUCAUUGGGUCUGGU .((((((((...((....))...))))))))((((((((...((.(((.(((((((....))))))).))).)).)))(((((((....))))))).)))))(((((.....)).))).. ( -50.70) >consensus ACUGCUGCAGGUGCGGAAGCUGAUGCAGCAGAGGUGGUUGUGGCUGCGGCUGGGGGCGACUUCCCGGAUGCUGCAAACGGCGUCGAAGCUGGCGUCACAUCUCAGCUCAUUGGGUCUGGU ..(((((((...((....))...)))))))(((.(((.((((((..(((((((((......)))).(((((((....)))))))..)))))..)))))).).)).)))............ (-52.13 = -52.13 + -0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:46:25 2006