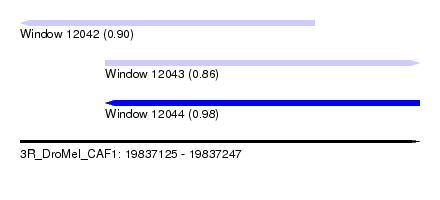
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 19,837,125 – 19,837,247 |
| Length | 122 |
| Max. P | 0.978865 |

| Location | 19,837,125 – 19,837,215 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 84.33 |
| Mean single sequence MFE | -35.82 |
| Consensus MFE | -26.60 |
| Energy contribution | -26.56 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.898279 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19837125 90 - 27905053 UUGCUUUUCGCUGUCCA--------CUUGGCACGGGGCCAAGGUUCGAGAAUUUGGCUCCCGUGU---GCGUUCAAUCCGUUCAUUUCGUUUGGCAGUGAC .......((((((.(((--------..(((((((((((((((.........)))))).)))))))---(((.......)))......))..))))))))). ( -33.00) >DroSec_CAF1 15404 92 - 1 UCGCUACUCGCUGGCCA--------CUUGGCACGGGGCCAAGGUUCGAGAAUUUGGCUCCCGUGCGC-GCGUUCAAUCCGUUCACUUCGUUUGGCAGUGAC .......((((((.(((--------..(((((((((((((((.........)))))).)))))))(.-(((.......))).)....))..))))))))). ( -36.00) >DroSim_CAF1 21756 92 - 1 UCGCUACUCGCUGGCCA--------CUUGGCACGGGGCCAAGGUUCGAGAAUUUGGCUCCCGUGCGC-GCGUUCAAUCCGUUCACUUCGUUUGGCAGUGAC .......((((((.(((--------..(((((((((((((((.........)))))).)))))))(.-(((.......))).)....))..))))))))). ( -36.00) >DroEre_CAF1 15515 85 - 1 UCGCCUCUAGCUGGCCA--------CUUGGCAUGGGGCCAAGGCUCGUGAAUUUGGCUC-CGUGCU---CGUCCAAUCCAUUC----CGUUUGGCAGUGAC ..(((.......)))((--------((.((((((((((((((.........))))))))-))))))---.(.((((.......----...))))))))).. ( -31.40) >DroYak_CAF1 15419 97 - 1 UCGCUGCUAGCUGGCCACUUGGCCACUUGGCAUGGGGCCAAGGCUCGUGAAUUUGGCUCCCGUGCUCUCCGUUCAAUCCAUUC----CGUUUGGCAGUGAC ((((((((((((((((....)))))...((((((((((((((.........)))))).)))))))).................----.).)))))))))). ( -42.70) >consensus UCGCUACUCGCUGGCCA________CUUGGCACGGGGCCAAGGUUCGAGAAUUUGGCUCCCGUGCGC_GCGUUCAAUCCGUUCA_UUCGUUUGGCAGUGAC .......((((((................(((((((((((((.........)))))).)))))))............(((..(.....)..))))))))). (-26.60 = -26.56 + -0.04)

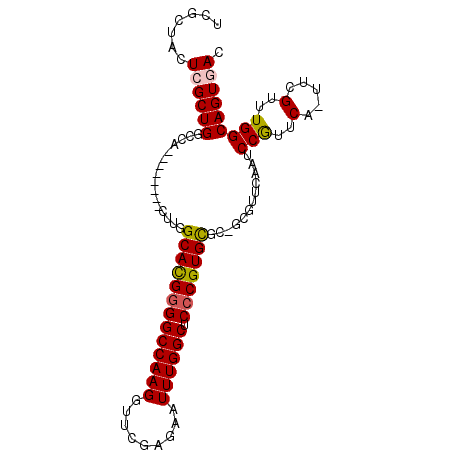
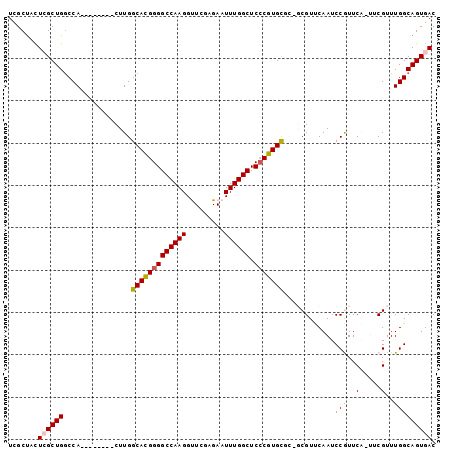
| Location | 19,837,151 – 19,837,247 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 76.54 |
| Mean single sequence MFE | -36.70 |
| Consensus MFE | -15.75 |
| Energy contribution | -17.00 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.43 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.861465 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19837151 96 + 27905053 UGAACGC---ACACGGGAGCCAAAUUCUCGAACCUUGGCCCCGUGCCAAG--------UGGACAGCGAAAAGCAAACUUCUGAUGGCCA---CCAGUCGAGUCGUUCAUC (((((((---.((((((.(((((...........)))))))))))....(--------(((.((..(((........)))...)).)))---).......).)))))).. ( -30.90) >DroSec_CAF1 15430 98 + 1 UGAACGC-GCGCACGGGAGCCAAAUUCUCGAACCUUGGCCCCGUGCCAAG--------UGGCCAGCGAGUAGCGAACUUCUGAUGGCCA---CCAGUCGAGUCGUUCAUC ((((((.-(((((((((.(((((...........))))))))))))...(--------((((((..((((.....))))....))))))---).......)))))))).. ( -43.10) >DroSim_CAF1 21782 98 + 1 UGAACGC-GCGCACGGGAGCCAAAUUCUCGAACCUUGGCCCCGUGCCAAG--------UGGCCAGCGAGUAGCGAACUUCUGAUGGCCA---CCAGUCGAGUCGUUCAUC ((((((.-(((((((((.(((((...........))))))))))))...(--------((((((..((((.....))))....))))))---).......)))))))).. ( -43.10) >DroEre_CAF1 15537 90 + 1 UGGACG---AGCACG-GAGCCAAAUUCACGAGCCUUGGCCCCAUGCCAAG--------UGGCCAGCUAGAGGCGAACUUCUGAUGGCCA---CCAG-----UCGUUCAUC ((((((---((((.(-(.(((((...........))))).)).)))...(--------((((((..((((((....)))))).))))))---)...-----))))))).. ( -38.90) >DroYak_CAF1 15441 105 + 1 UGAACGGAGAGCACGGGAGCCAAAUUCACGAGCCUUGGCCCCAUGCCAAGUGGCCAAGUGGCCAGCUAGCAGCGAACUUUUGAUGGCCA---CCAGU--AGUCGUUCAUC (((((((...((..((..((((...(((.(((.((((((.....))))))(((((....)))))((.....))...))).)))))))..---)).))--..))))))).. ( -38.10) >DroPer_CAF1 30671 86 + 1 UGCAGG------------CCAAAAUCCACGUUCCUUGGCCACUCGCCAAG--------UGC---GCGACAAAUGAACUUUUGAUGGUCAAACCCAGUGGGGCAGU-CACC (((.((------------(((.......(((.(((((((.....))))))--------.).---))).((((......)))).)))))...(((...))))))..-.... ( -26.10) >consensus UGAACGC___GCACGGGAGCCAAAUUCACGAACCUUGGCCCCGUGCCAAG________UGGCCAGCGAGAAGCGAACUUCUGAUGGCCA___CCAGUCGAGUCGUUCAUC ((((((...........................((((((.....))))))........((((((((.....))...(....).)))))).............)))))).. (-15.75 = -17.00 + 1.25)

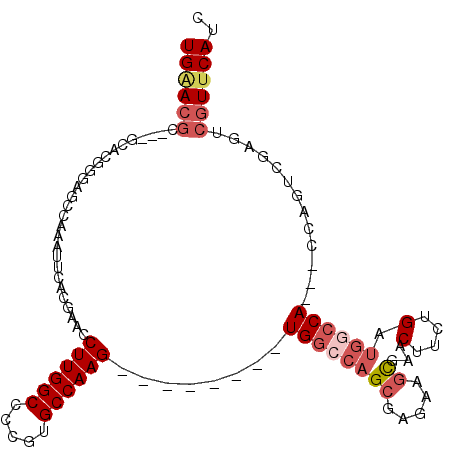
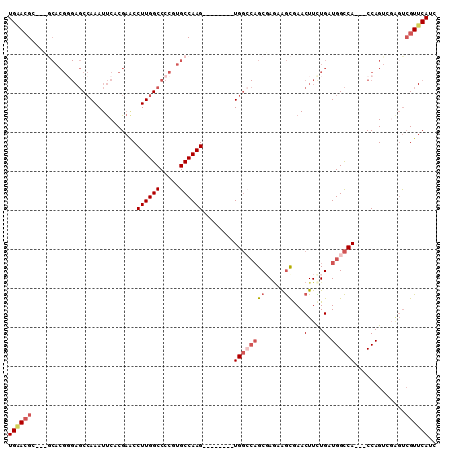
| Location | 19,837,151 – 19,837,247 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 76.54 |
| Mean single sequence MFE | -40.53 |
| Consensus MFE | -21.76 |
| Energy contribution | -24.93 |
| Covariance contribution | 3.17 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.54 |
| SVM decision value | 1.82 |
| SVM RNA-class probability | 0.978865 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19837151 96 - 27905053 GAUGAACGACUCGACUGG---UGGCCAUCAGAAGUUUGCUUUUCGCUGUCCA--------CUUGGCACGGGGCCAAGGUUCGAGAAUUUGGCUCCCGUGU---GCGUUCA ..((((((......(.((---(((.((.((((((....))))).).)).)))--------)).)(((((((((((((.........)))))).)))))))---.)))))) ( -38.00) >DroSec_CAF1 15430 98 - 1 GAUGAACGACUCGACUGG---UGGCCAUCAGAAGUUCGCUACUCGCUGGCCA--------CUUGGCACGGGGCCAAGGUUCGAGAAUUUGGCUCCCGUGCGC-GCGUUCA ..((((((.(.(..(.((---((((((.(((.((....)).)).).))))))--------)).)(((((((((((((.........)))))).)))))))).-))))))) ( -45.60) >DroSim_CAF1 21782 98 - 1 GAUGAACGACUCGACUGG---UGGCCAUCAGAAGUUCGCUACUCGCUGGCCA--------CUUGGCACGGGGCCAAGGUUCGAGAAUUUGGCUCCCGUGCGC-GCGUUCA ..((((((.(.(..(.((---((((((.(((.((....)).)).).))))))--------)).)(((((((((((((.........)))))).)))))))).-))))))) ( -45.60) >DroEre_CAF1 15537 90 - 1 GAUGAACGA-----CUGG---UGGCCAUCAGAAGUUCGCCUCUAGCUGGCCA--------CUUGGCAUGGGGCCAAGGCUCGUGAAUUUGGCUC-CGUGCU---CGUCCA (((((....-----(.((---((((((.((((.(....).))).).))))))--------)).)(((((((((((((.........))))))))-))))))---)))).. ( -41.80) >DroYak_CAF1 15441 105 - 1 GAUGAACGACU--ACUGG---UGGCCAUCAAAAGUUCGCUGCUAGCUGGCCACUUGGCCACUUGGCAUGGGGCCAAGGCUCGUGAAUUUGGCUCCCGUGCUCUCCGUUCA ..((((((...--.(.((---((((((......((..((.....))..))....)))))))).)(((((((((((((.........)))))).)))))))....)))))) ( -44.40) >DroPer_CAF1 30671 86 - 1 GGUG-ACUGCCCCACUGGGUUUGACCAUCAAAAGUUCAUUUGUCGC---GCA--------CUUGGCGAGUGGCCAAGGAACGUGGAUUUUGG------------CCUGCA (((.-(..((((....)))).).)))......((..((....((((---(..--------((((((.....))))))...)))))....)).------------.))... ( -27.80) >consensus GAUGAACGACUCGACUGG___UGGCCAUCAGAAGUUCGCUUCUCGCUGGCCA________CUUGGCACGGGGCCAAGGUUCGAGAAUUUGGCUCCCGUGC___GCGUUCA ..((((((.............((((((.(((.((....)).)).).))))))............(((((((((((((.........)))))).)))))))....)))))) (-21.76 = -24.93 + 3.17)

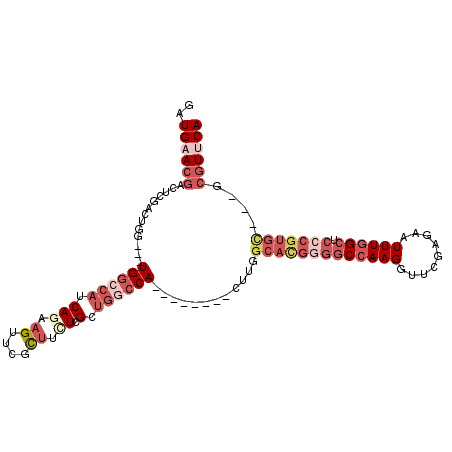
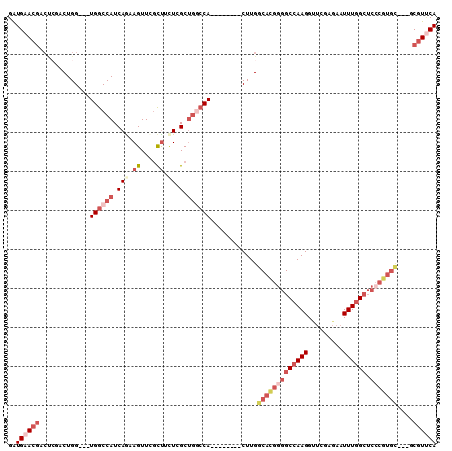
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:46:18 2006