
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 19,836,118 – 19,836,250 |
| Length | 132 |
| Max. P | 0.985149 |

| Location | 19,836,118 – 19,836,231 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 86.06 |
| Mean single sequence MFE | -23.62 |
| Consensus MFE | -21.50 |
| Energy contribution | -21.00 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.00 |
| SVM RNA-class probability | 0.985149 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19836118 113 + 27905053 CAUAGUUGAUACCACGAGUGUUAUUGCCAUGAAAAAUUAAUGAGCCAAACACUCCGAUUAAAACACUCAUAAAACAUUUAUGAGUACUUAAC-CGGCAGAACACAUAUUCA-----GUA .................(((((.(((((........((((((((.......)))..)))))...((((((((.....)))))))).......-.)))))))))).......-----... ( -22.10) >DroSec_CAF1 14371 118 + 1 CGUAGUUGAUACCACGAGUGUUAUUGCCAUGAAAAAUUAAUGAGCCGAACACUCCGAUUAAAACACUCAUAAAACAUUUAUGAGUACUUAAC-CGGCAGAACGUACAUAUAUAUAUGUA (((..(((.......(((((((...(((((.........))).))..)))))))((.((((...((((((((.....))))))))..)))).-)).))).)))((((((....)))))) ( -25.20) >DroEre_CAF1 14468 114 + 1 CGUAGUUAAUACCACGAGUGUUAUUGCCAUGCAAAAUGAAUGAGCCGAACACUCCGAUUUAAACACUUAUAAAACAUUUAUGAGUACUUAAC-CAGCAGAACAUAUACAUGCCUA---- ....(((((......(((((((...(((((.(.....).))).))..)))))))..........((((((((.....))))))))..)))))-..(((...........)))...---- ( -20.30) >DroYak_CAF1 14434 119 + 1 CGCAGUUGAUACUGCGAGUGUUAUUGCCAUGAAAAAUUAAUGAGCCGAACACUCCGAUUAAAGCACUCAUAAAACAUUUAUGAGUACUCAACCCAGCAGAACACAUACAUACAUAUUCA .(((((....)))))(((((((...(((((.........))).))..))))))).......((.((((((((.....)))))))).))............................... ( -26.90) >consensus CGUAGUUGAUACCACGAGUGUUAUUGCCAUGAAAAAUUAAUGAGCCGAACACUCCGAUUAAAACACUCAUAAAACAUUUAUGAGUACUUAAC_CAGCAGAACACAUACAUACAUA_GUA .((.(((((......(((((((...(((((.........))).))..)))))))..........((((((((.....))))))))..)))))...))...................... (-21.50 = -21.00 + -0.50)

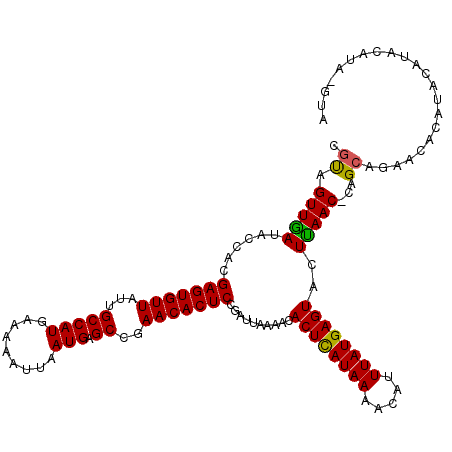

| Location | 19,836,118 – 19,836,231 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 86.06 |
| Mean single sequence MFE | -31.10 |
| Consensus MFE | -26.12 |
| Energy contribution | -24.62 |
| Covariance contribution | -1.50 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.78 |
| SVM RNA-class probability | 0.977126 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19836118 113 - 27905053 UAC-----UGAAUAUGUGUUCUGCCG-GUUAAGUACUCAUAAAUGUUUUAUGAGUGUUUUAAUCGGAGUGUUUGGCUCAUUAAUUUUUCAUGGCAAUAACACUCGUGGUAUCAACUAUG ...-----.......(((((.(((((-(((((((((((((((.....))))))))).))))))).((((.....)))).............))))..))))).((((((....)))))) ( -28.60) >DroSec_CAF1 14371 118 - 1 UACAUAUAUAUAUGUACGUUCUGCCG-GUUAAGUACUCAUAAAUGUUUUAUGAGUGUUUUAAUCGGAGUGUUCGGCUCAUUAAUUUUUCAUGGCAAUAACACUCGUGGUAUCAACUACG ((((((....))))))........((-(((((((((((((((.....))))))))).))))))))(((((((..((.(((.........)))))...)))))))(((((....))))). ( -31.90) >DroEre_CAF1 14468 114 - 1 ----UAGGCAUGUAUAUGUUCUGCUG-GUUAAGUACUCAUAAAUGUUUUAUAAGUGUUUAAAUCGGAGUGUUCGGCUCAUUCAUUUUGCAUGGCAAUAACACUCGUGGUAUUAACUACG ----..(.((((((.(((....((((-(.....(((((.((((..((.....))..)))).....))))).))))).....)))..)))))).).........((((((....)))))) ( -25.00) >DroYak_CAF1 14434 119 - 1 UGAAUAUGUAUGUAUGUGUUCUGCUGGGUUGAGUACUCAUAAAUGUUUUAUGAGUGCUUUAAUCGGAGUGUUCGGCUCAUUAAUUUUUCAUGGCAAUAACACUCGCAGUAUCAACUGCG .((((((((....)))))))).....((((((((((((((((.....)))))))))).)))))).(((((((..((.(((.........)))))...)))))))(((((....))))). ( -38.90) >consensus UAC_UAUGUAUAUAUAUGUUCUGCCG_GUUAAGUACUCAUAAAUGUUUUAUGAGUGUUUUAAUCGGAGUGUUCGGCUCAUUAAUUUUUCAUGGCAAUAACACUCGUGGUAUCAACUACG ........................((....((((((((((((.....))))))))))))....))(((((((..((.(((.(.....).)))))...)))))))(((((....))))). (-26.12 = -24.62 + -1.50)


| Location | 19,836,158 – 19,836,250 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 78.14 |
| Mean single sequence MFE | -25.30 |
| Consensus MFE | -14.90 |
| Energy contribution | -14.14 |
| Covariance contribution | -0.75 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.811350 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19836158 92 - 27905053 GAGGAUUGUAAGGAAUAU------GUAC-----UGAAUAUGUGUUCUGCCG-GUUAAGUACUCAUAAAUGUUUUAUGAGUGUUUUAAUCGGAGUGUUUGGCUCA (((........(((((((------(((.-----....)))))))))).(((-(((((((((((((((.....))))))))).))))))))).........))). ( -26.00) >DroSec_CAF1 14411 97 - 1 GAGGACUGUAAGGAAUAU------AUACAUAUAUAUAUGUACGUUCUGCCG-GUUAAGUACUCAUAAAUGUUUUAUGAGUGUUUUAAUCGGAGUGUUCGGCUCA (((.(((....(((((..------.((((((....)))))).))))).(((-(((((((((((((((.....))))))))).)))))))))))).)))...... ( -27.60) >DroEre_CAF1 14508 98 - 1 AAGGACGUAAUGGAAUC-GUCGAAA----UAGGCAUGUAUAUGUUCUGCUG-GUUAAGUACUCAUAAAUGUUUUAUAAGUGUUUAAAUCGGAGUGUUCGGCUCA .((((((((.....((.-(((....----..)))))...))))))))((((-(.....(((((.((((..((.....))..)))).....))))).)))))... ( -17.80) >DroYak_CAF1 14474 103 - 1 AAGGACUGCAAGGAAUC-GUCGAAAUGAAUAUGUAUGUAUGUGUUCUGCUGGGUUGAGUACUCAUAAAUGUUUUAUGAGUGCUUUAAUCGGAGUGUUCGGCUCA .................-((((((..((((((((....))))))))..((((...((((((((((((.....))))))))))))...))))....))))))... ( -29.80) >consensus AAGGACUGUAAGGAAUA_______AUAC_UAUGUAUAUAUAUGUUCUGCCG_GUUAAGUACUCAUAAAUGUUUUAUGAGUGUUUUAAUCGGAGUGUUCGGCUCA ..((((.....(((((..........................))))).(((....((((((((((((.....))))))))))))....)))...))))...... (-14.90 = -14.14 + -0.75)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:46:15 2006