
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 19,830,838 – 19,830,954 |
| Length | 116 |
| Max. P | 0.776094 |

| Location | 19,830,838 – 19,830,954 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.09 |
| Mean single sequence MFE | -28.56 |
| Consensus MFE | -17.71 |
| Energy contribution | -18.03 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.776094 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19830838 116 + 27905053 UGGAAAUGCCUUAU----AGAUUAGCUUAUGCUAAUUUCUGGGAAAUUCCUGAAUAGGCAUGGGAAGAAGAAAUUAGAACAAGUUAAUCCUAAGAGAUAUCUGACAGAAUCUAUGCAAAU ......(((((((.----.(((((((((...(((((((((......(((((..........)))))..)))))))))...))))))))).))))((((.((.....))))))..)))... ( -27.80) >DroSec_CAF1 9067 116 + 1 UGGAAAUGCCUUAU----AGAUUAGCUUAUGCUAAUUUCUGGGAAAUUCCUAAAAAGGCAUGGCUAGAAGGAACUAGAACAAGGUAAUCCUAAGAGAUAUCUGACUGAAUCUAGUUAAAU (((..(((((((..----((((((((....)))))))).((((.....))))..)))))))..))).....(((((((...(((....))).(((....))).......))))))).... ( -32.60) >DroSim_CAF1 13790 116 + 1 UGGAAAUGCCUUAU----AGAUUAGCUUAUGCUAAUUUCUGGGAAAUUCCUAAAAAGGCAUGGCUAGAAGGAACUAGAACAAGGUAAUCCUAAGAUAUAUCUCACUGAAUCUAGUUAAAU (((..(((((((..----((((((((....)))))))).((((.....))))..)))))))..))).....(((((((...(((....)))........((.....)).))))))).... ( -31.50) >DroEre_CAF1 9132 105 + 1 UGGAAAUGCCUUAU----AGAUUAGCUUAUGCUAAUUUCUGCGAAAUUCCUUAAAAG-----GGUAGAAGGAACUAGAACAAGGUAUUCCUAAGAGAUAUCUGACUGAAUCUAG------ .((((.((((((.(----((.(((((....)))))(((((((......(((....))-----))))))))...)))....))))))))))....((((.((.....))))))..------ ( -24.10) >DroYak_CAF1 9081 116 + 1 UGGAAAUGCCUUAUACAUAGAUUAGCUUAUGCUAAUUUUUGGGAAAUUCCGGAAAAGGCAUGGAUGGAAGGCACUAGAACAAGGUAAUCCUGAAAGAUAUCAGACUGAAUCUAA----AU .....(((((((...((.((((((((....)))))))).))((.....))....)))))))((((...((((.((......))))....((((......)))).))..))))..----.. ( -26.80) >consensus UGGAAAUGCCUUAU____AGAUUAGCUUAUGCUAAUUUCUGGGAAAUUCCUAAAAAGGCAUGGCUAGAAGGAACUAGAACAAGGUAAUCCUAAGAGAUAUCUGACUGAAUCUAG__AAAU .(((..((((((.......(((((((....)))))))(((((....(((.................)))....)))))..)))))).)))....((((.((.....))))))........ (-17.71 = -18.03 + 0.32)



| Location | 19,830,838 – 19,830,954 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.09 |
| Mean single sequence MFE | -22.39 |
| Consensus MFE | -14.15 |
| Energy contribution | -14.43 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.63 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.512054 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19830838 116 - 27905053 AUUUGCAUAGAUUCUGUCAGAUAUCUCUUAGGAUUAACUUGUUCUAAUUUCUUCUUCCCAUGCCUAUUCAGGAAUUUCCCAGAAAUUAGCAUAAGCUAAUCU----AUAAGGCAUUUCCA ...(((..((((((.....)).))))((((((((((.(((((.(((((((((..........(((....)))........))))))))).))))).))))))----.)))))))...... ( -25.47) >DroSec_CAF1 9067 116 - 1 AUUUAACUAGAUUCAGUCAGAUAUCUCUUAGGAUUACCUUGUUCUAGUUCCUUCUAGCCAUGCCUUUUUAGGAAUUUCCCAGAAAUUAGCAUAAGCUAAUCU----AUAAGGCAUUUCCA ....(((((((..(((......((((....))))....))).)))))))..........(((((((....((......))(((..(((((....))))))))----..)))))))..... ( -21.60) >DroSim_CAF1 13790 116 - 1 AUUUAACUAGAUUCAGUGAGAUAUAUCUUAGGAUUACCUUGUUCUAGUUCCUUCUAGCCAUGCCUUUUUAGGAAUUUCCCAGAAAUUAGCAUAAGCUAAUCU----AUAAGGCAUUUCCA ....(((((((..((((((((....)))))((....))))).)))))))..........(((((((....((......))(((..(((((....))))))))----..)))))))..... ( -23.40) >DroEre_CAF1 9132 105 - 1 ------CUAGAUUCAGUCAGAUAUCUCUUAGGAAUACCUUGUUCUAGUUCCUUCUACC-----CUUUUAAGGAAUUUCGCAGAAAUUAGCAUAAGCUAAUCU----AUAAGGCAUUUCCA ------..((((((.....)).))))....((((..(((((((((((((((((.....-----.....))))))))....))))((((((....))))))..----.)))))...)))). ( -21.10) >DroYak_CAF1 9081 116 - 1 AU----UUAGAUUCAGUCUGAUAUCUUUCAGGAUUACCUUGUUCUAGUGCCUUCCAUCCAUGCCUUUUCCGGAAUUUCCCAAAAAUUAGCAUAAGCUAAUCUAUGUAUAAGGCAUUUCCA ..----.((((..((((((((......)))))......))).))))(((((((..((.((((........((......))....((((((....)))))).)))).)))))))))..... ( -20.40) >consensus AUUU__CUAGAUUCAGUCAGAUAUCUCUUAGGAUUACCUUGUUCUAGUUCCUUCUACCCAUGCCUUUUCAGGAAUUUCCCAGAAAUUAGCAUAAGCUAAUCU____AUAAGGCAUUUCCA ..............................(((...((((((...(((((((.................)))))))........((((((....))))))......))))))....))). (-14.15 = -14.43 + 0.28)

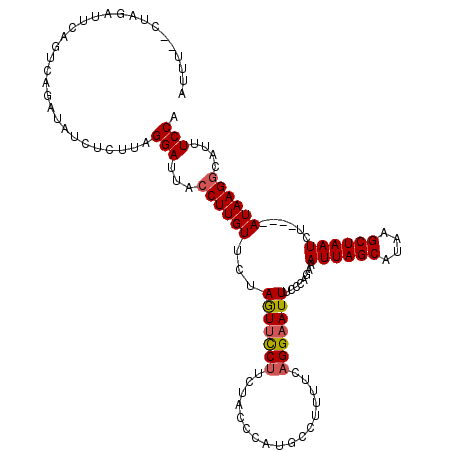
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:46:11 2006