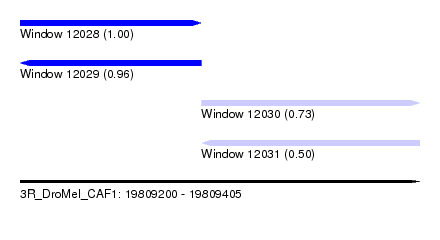
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 19,809,200 – 19,809,405 |
| Length | 205 |
| Max. P | 0.996732 |

| Location | 19,809,200 – 19,809,293 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 85.61 |
| Mean single sequence MFE | -46.73 |
| Consensus MFE | -43.18 |
| Energy contribution | -43.77 |
| Covariance contribution | 0.58 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.26 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.74 |
| SVM RNA-class probability | 0.996732 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
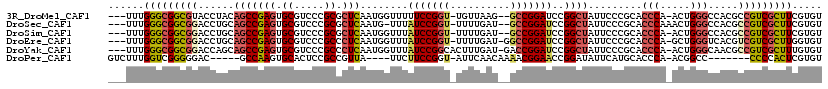
>3R_DroMel_CAF1 19809200 93 + 27905053 GCAGUUGAGUAUCGCCG----------GCCAGGGAAGGGUCGAGUGCGGAUCCCUUGCCCCAGCGGCUAUUCGCCUGCGCACUCGGUCCAUCCCUGGCUCACU ................(----------((((((((.((..(((((((((..........)).(((((.....))).)))))))))..)).))))))))).... ( -46.90) >DroSec_CAF1 39661 93 + 1 GCAGUUGAGUAUCGCCG----------GCCAGGGAAGGGUCGAGUGCGGAUCCCUUGCCCCAGCGGCUAUUCGCCUGCGCACUCGGCCCAUCCCUGGCACACU .................----------((((((((.(((((((((((((..........)).(((((.....))).))))))))))))).))))))))..... ( -49.80) >DroSim_CAF1 39739 93 + 1 GCAGUUGAGUAUCGCCG----------GCCAGGGAAGGGUCGAAUGCGGAUCCCUUGCCCCAGCGGCUAUUCGCCUGCGCACUCAACCCAUCCCUGGCACACU .................----------((((((((.((((.((.(((((..........)).(((((.....))).))))).)).)))).))))))))..... ( -38.50) >DroEre_CAF1 40296 93 + 1 GCAGUUGAGUAUCGCGG----------GCCAGGGAAGGGUCGAGUGCGGAUCCCUUGCCCCAGCGGCUAUUCGCCUGCGCACUCGGCCCAUCCCUGGCACACU ((.((.....)).))..----------((((((((.(((((((((((((..........)).(((((.....))).))))))))))))).))))))))..... ( -50.40) >DroYak_CAF1 40423 93 + 1 GCAGUUGAGCAUCGCGG----------GCCAGGGAAGGGUCGAGUGCGGAUCCCUUGCCCCAGCGGCUAUUCGCCUGCGCACUCGGCCCAUCCCUGGCACACU ((......)).......----------((((((((.(((((((((((((..........)).(((((.....))).))))))))))))).))))))))..... ( -51.00) >DroAna_CAF1 41532 95 + 1 --------GAAUCGCGACUAGGAUGCCUCCAUCGGUGGGUCGAGUGCGAAUCUCCCGCGCCUGCGGAUAUUCGCCUGCGCACUCGGCCCAUCCCUGGUACACG --------....((.((((((((((....))))((((((((((((((((((...((((....))))..))))(....))))))))))))))).))))).).)) ( -43.80) >consensus GCAGUUGAGUAUCGCCG__________GCCAGGGAAGGGUCGAGUGCGGAUCCCUUGCCCCAGCGGCUAUUCGCCUGCGCACUCGGCCCAUCCCUGGCACACU ...........................((((((((.((((((((((((........((....))(((.....)))..)))))))))))).))))))))..... (-43.18 = -43.77 + 0.58)


| Location | 19,809,200 – 19,809,293 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 85.61 |
| Mean single sequence MFE | -47.38 |
| Consensus MFE | -40.00 |
| Energy contribution | -40.58 |
| Covariance contribution | 0.59 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.98 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.963004 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19809200 93 - 27905053 AGUGAGCCAGGGAUGGACCGAGUGCGCAGGCGAAUAGCCGCUGGGGCAAGGGAUCCGCACUCGACCCUUCCCUGGC----------CGGCGAUACUCAACUGC .((..((((((((.((..(((((((((.(((.....))))).(((........))))))))))..)).))))))))----------..))............. ( -44.90) >DroSec_CAF1 39661 93 - 1 AGUGUGCCAGGGAUGGGCCGAGUGCGCAGGCGAAUAGCCGCUGGGGCAAGGGAUCCGCACUCGACCCUUCCCUGGC----------CGGCGAUACUCAACUGC .((..((((((((.(((.(((((((((.(((.....))))).(((........)))))))))).))).))))))))----------..))............. ( -48.00) >DroSim_CAF1 39739 93 - 1 AGUGUGCCAGGGAUGGGUUGAGUGCGCAGGCGAAUAGCCGCUGGGGCAAGGGAUCCGCAUUCGACCCUUCCCUGGC----------CGGCGAUACUCAACUGC .((..((((((((.(((((((((((((.(((.....))))).(((........)))))))))))))).))))))))----------..))............. ( -48.60) >DroEre_CAF1 40296 93 - 1 AGUGUGCCAGGGAUGGGCCGAGUGCGCAGGCGAAUAGCCGCUGGGGCAAGGGAUCCGCACUCGACCCUUCCCUGGC----------CCGCGAUACUCAACUGC .(((.((((((((.(((.(((((((((.(((.....))))).(((........)))))))))).))).))))))))----------.)))............. ( -50.90) >DroYak_CAF1 40423 93 - 1 AGUGUGCCAGGGAUGGGCCGAGUGCGCAGGCGAAUAGCCGCUGGGGCAAGGGAUCCGCACUCGACCCUUCCCUGGC----------CCGCGAUGCUCAACUGC .(((.((((((((.(((.(((((((((.(((.....))))).(((........)))))))))).))).))))))))----------.)))............. ( -50.90) >DroAna_CAF1 41532 95 - 1 CGUGUACCAGGGAUGGGCCGAGUGCGCAGGCGAAUAUCCGCAGGCGCGGGAGAUUCGCACUCGACCCACCGAUGGAGGCAUCCUAGUCGCGAUUC-------- ((((.((.((((.((((.((((((((....)((((.(((((....)))))..))))))))))).))))))((((....)))))).))))))....-------- ( -41.00) >consensus AGUGUGCCAGGGAUGGGCCGAGUGCGCAGGCGAAUAGCCGCUGGGGCAAGGGAUCCGCACUCGACCCUUCCCUGGC__________CCGCGAUACUCAACUGC .....((((((((.(((.(((((((((.(((.....))))).((..(....)..))))))))).))).))))))))........................... (-40.00 = -40.58 + 0.59)


| Location | 19,809,293 – 19,809,405 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 80.30 |
| Mean single sequence MFE | -44.56 |
| Consensus MFE | -29.39 |
| Energy contribution | -30.58 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.725520 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19809293 112 + 27905053 ACACGAAGCGACGGCGUGGCCCAGU-UGGGUGCGGGAAUAGCCGGAUCCGGC--CUUAACA-ACCGGAAAAACCAUUGAGCGCGGGACGCACUCGGCUGUAGGUACGCCGCCCAAA--- .......(.(.(((((((.((((((-((((((((((.....))...(((.((--(((((..-...((.....)).))))).)).)))))))))))))))..))))))))).))...--- ( -44.80) >DroSec_CAF1 39754 112 + 1 ACACGAAGCGACGGCGUGGCCCAGUUUGGGUGCGGGAAUAGCCGGAUCCGGC--AUCAAAA-ACCGGAUAAA-CAUUGAGCGCGGGACGCACUCGGCUGCAGGUCCGCCGCCCAAA--- .((((.........))))......((((((((((((..((((((.((((((.--.......-.))))))...-)...(((.(((...))).))))))))....)))).))))))))--- ( -42.90) >DroSim_CAF1 39832 112 + 1 ACACGAAGCGACGGCGUGGCCCAGU-UGGGUGCGGGAAUAGCCGGAUCCGGC--AUCAAAA-ACCGGAUAAACCAUUGAGCGCGGGACGCACUCGGCUGCAGGUCCGCCGCCCAAA--- .......(.(.(((((.((((((((-((((((((......((((....))))--.......-.(((................)))..))))))))))))..))))))))).))...--- ( -46.19) >DroEre_CAF1 40389 113 + 1 ACACCAAGCGACGACGUGACCCAGC-UGGGUGCGGGAAUAGCCGGAUCCGGCC-AUCAAAA-ACCGGAUAAACCAUUGAGGGCGGGACGCACUCGGCUGCAGGUCCGCCGCCCAAA--- .......(((..(.((.((((((((-((((((((((.....))...(((.(((-.((((..-...((.....)).)))).))).)))))))))))))))..)))))))))).....--- ( -45.80) >DroYak_CAF1 40516 114 + 1 ACACAAAGCGACGGCGUUGCCCAGU-UGGGUGCGGGAAUAGCCGGAUCCGGUC-AUCAAAGUGCCGGAUAAACCAUUGAGGGCGGGACGCACUCGGCUGCUGGUCCGCCGCCCAAA--- .......(((((...)))))....(-((((((((((..(((((((((((((.(-((....)))))))))...))...(((.(((...))).))))))))....)))).))))))).--- ( -46.00) >DroPer_CAF1 44033 101 + 1 ACACGAGUGGGG-------GGCCGU-UGGGUGCAUGAAUAUCCGGUUCCGUUUUGUUGAAU-ACCGGAAGAA----UAACGGCGGAGUGCACUUGGC-----GUCCCCCGACCAAAGAC ....(.((.(((-------((.(((-..(((((((.....((((...(((((..((.....-))(....)..----.))))))))))))))))..))-----).))))).)))...... ( -41.70) >consensus ACACGAAGCGACGGCGUGGCCCAGU_UGGGUGCGGGAAUAGCCGGAUCCGGC__AUCAAAA_ACCGGAUAAACCAUUGAGCGCGGGACGCACUCGGCUGCAGGUCCGCCGCCCAAA___ ..........................((((((((((..(((((..((((((............))))))........(((.(((...))).))))))))....)))).))))))..... (-29.39 = -30.58 + 1.20)

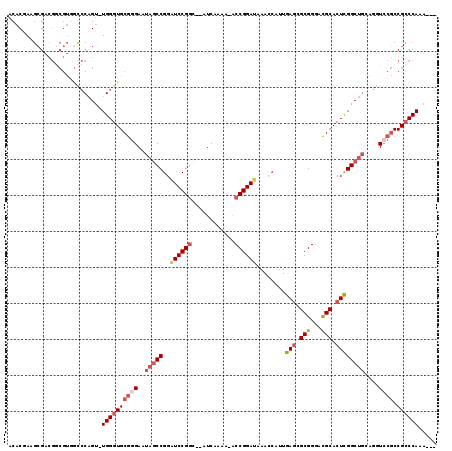
| Location | 19,809,293 – 19,809,405 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 80.30 |
| Mean single sequence MFE | -42.73 |
| Consensus MFE | -30.91 |
| Energy contribution | -31.38 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.07 |
| Structure conservation index | 0.72 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
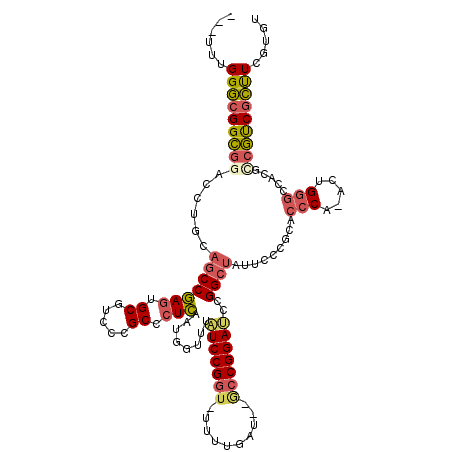
>3R_DroMel_CAF1 19809293 112 - 27905053 ---UUUGGGCGGCGUACCUACAGCCGAGUGCGUCCCGCGCUCAAUGGUUUUUCCGGU-UGUUAAG--GCCGGAUCCGGCUAUUCCCGCACCCA-ACUGGGCCACGCCGUCGCUUCGUGU ---....((((((((......(((((((((((...)))))))...((....((((((-(.....)--)))))).)))))).........(((.-...)))..))))))))......... ( -48.20) >DroSec_CAF1 39754 112 - 1 ---UUUGGGCGGCGGACCUGCAGCCGAGUGCGUCCCGCGCUCAAUG-UUUAUCCGGU-UUUUGAU--GCCGGAUCCGGCUAUUCCCGCACCCAAACUGGGCCACGCCGUCGCUUCGUGU ---...(((((((((...((((((((((((((...)))))))....-...(((((((-.......--)))))))..))))......)))(((.....))).....)))))))))..... ( -45.40) >DroSim_CAF1 39832 112 - 1 ---UUUGGGCGGCGGACCUGCAGCCGAGUGCGUCCCGCGCUCAAUGGUUUAUCCGGU-UUUUGAU--GCCGGAUCCGGCUAUUCCCGCACCCA-ACUGGGCCACGCCGUCGCUUCGUGU ---...(((((((((...((((((((((((((...)))))))...((...(((((((-.......--)))))))))))))......)))(((.-...))).....)))))))))..... ( -46.90) >DroEre_CAF1 40389 113 - 1 ---UUUGGGCGGCGGACCUGCAGCCGAGUGCGUCCCGCCCUCAAUGGUUUAUCCGGU-UUUUGAU-GGCCGGAUCCGGCUAUUCCCGCACCCA-GCUGGGUCACGUCGUCGCUUGGUGU ---....(((((((((((..((((.(.(((((..(((.((.....))......))).-....(((-(((((....))))))))..))))).).-)))))))).)))))))......... ( -45.70) >DroYak_CAF1 40516 114 - 1 ---UUUGGGCGGCGGACCAGCAGCCGAGUGCGUCCCGCCCUCAAUGGUUUAUCCGGCACUUUGAU-GACCGGAUCCGGCUAUUCCCGCACCCA-ACUGGGCAACGCCGUCGCUUUGUGU ---...(((((((((....(((((((((.(((...))).)))...((...((((((((......)-).))))))))))))..........((.-...))))....)))))))))..... ( -41.30) >DroPer_CAF1 44033 101 - 1 GUCUUUGGUCGGGGGAC-----GCCAAGUGCACUCCGCCGUUA----UUCUUCCGGU-AUUCAACAAAACGGAACCGGAUAUUCAUGCACCCA-ACGGCC-------CCCCACUCGUGU ......(((.(((((..-----((.....)).....((((((.----....((((((-.(((........))))))))).............)-))))))-------)))))))..... ( -28.87) >consensus ___UUUGGGCGGCGGACCUGCAGCCGAGUGCGUCCCGCCCUCAAUGGUUUAUCCGGU_UUUUGAU__GCCGGAUCCGGCUAUUCCCGCACCCA_ACUGGGCCACGCCGUCGCUUCGUGU ......(((((((((......(((((((.((.....)).)))........(((((((..........)))))))..)))).........(((.....))).....)))))))))..... (-30.91 = -31.38 + 0.48)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:46:06 2006