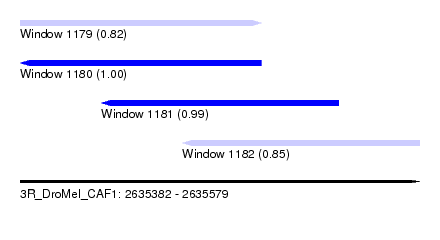
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 2,635,382 – 2,635,579 |
| Length | 197 |
| Max. P | 0.999919 |

| Location | 2,635,382 – 2,635,501 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.99 |
| Mean single sequence MFE | -36.02 |
| Consensus MFE | -34.02 |
| Energy contribution | -34.42 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.822465 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2635382 119 + 27905053 AUGUUAAUAAACAUAUACAAUGUGUGUGUUCAAGGCGGGGUAAUUUAGGCAGCGCCGCAAUUACAUGCGCCUCGGGUAAAUUAUAUUUAUCGUGCCACACGUGAUGCGGCUGUGGCUAG- .........(((((((((...)))))))))(.(((((.(((((((..(((...)))..)))))).).))))).)(((((((...)))))))..((((((((.....))..))))))...- ( -35.20) >DroSec_CAF1 35698 119 + 1 AUAUUAAUAAACAUAUACAAUGUGUGUGUUCAAGGCGGGGUAAUUUAGGCAGCGCCGCAAUUACAUGCGCCUCGGGUAAAUUAUAUUUAUAGUGCCACACGUGAUGCGGCGGUGGCUAG- ..........((.((((.((((((..((..(.(((((.(((((((..(((...)))..)))))).).))))).)..))...))))))))))))(((((.(((......))))))))...- ( -35.70) >DroSim_CAF1 23914 119 + 1 AUGUUAAUAAACAUAUACAAUGUGUGUGUUCAAGGCGGGGUAAUUUAGGCAGCGCCGCAAUUACAUGCGCCUCGGGUAAAUUAUAUUUAUCGUGCCACACGUGAUGCGGCGGUGGCUAG- .........(((((((((...)))))))))(.(((((.(((((((..(((...)))..)))))).).))))).)(((((((...)))))))..(((((.(((......))))))))...- ( -36.70) >DroEre_CAF1 26398 119 + 1 AUGUUAAUAAUCAUAUACAAUGUGUGUGUUCAAGGCGGGGUAAUUUAGGCAGCGCCGCAAUUACAUGCGCCUCGGGUAAAUUAUAUUUAUCGUGCCACACGUGAUGCGGCGGUGGCUAG- ...........(((((((...)))))))..(.(((((.(((((((..(((...)))..)))))).).))))).)(((((((...)))))))..(((((.(((......))))))))...- ( -34.40) >DroYak_CAF1 17355 120 + 1 AUGUUAAUAAACAUAUACAAUGUGUGUGUUCAAGGCGGGGUAAUUUAGGCAGCGCCGCAAUUACAUGCGCCUCGGGUAAAUUAUAUUUAUCGUGCCACACGUGAUGCGGCGGUGGCAAGC .........(((((((((...)))))))))(.(((((.(((((((..(((...)))..)))))).).))))).)(((((((...))))))).((((((.(((......)))))))))... ( -38.10) >consensus AUGUUAAUAAACAUAUACAAUGUGUGUGUUCAAGGCGGGGUAAUUUAGGCAGCGCCGCAAUUACAUGCGCCUCGGGUAAAUUAUAUUUAUCGUGCCACACGUGAUGCGGCGGUGGCUAG_ .........(((((((((...)))))))))(.(((((.(((((((..(((...)))..)))))).).))))).)(((((((...)))))))..(((((.((.....))...))))).... (-34.02 = -34.42 + 0.40)

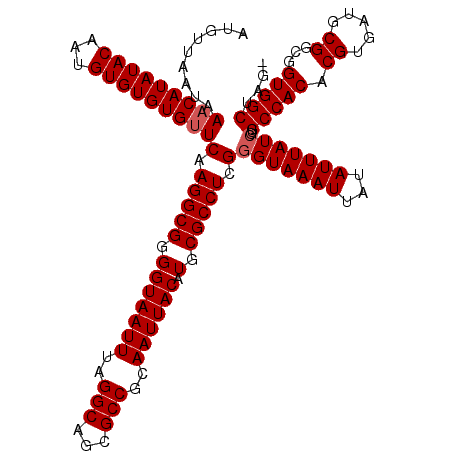
| Location | 2,635,382 – 2,635,501 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.99 |
| Mean single sequence MFE | -33.68 |
| Consensus MFE | -33.16 |
| Energy contribution | -33.56 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.87 |
| Structure conservation index | 0.98 |
| SVM decision value | 4.55 |
| SVM RNA-class probability | 0.999919 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2635382 119 - 27905053 -CUAGCCACAGCCGCAUCACGUGUGGCACGAUAAAUAUAAUUUACCCGAGGCGCAUGUAAUUGCGGCGCUGCCUAAAUUACCCCGCCUUGAACACACACAUUGUAUAUGUUUAUUAACAU -.........(((((((...)))))))..((((((((((.......(((((((...((((((..(((...)))..))))))..)))))))...(((.....))).))))))))))..... ( -34.40) >DroSec_CAF1 35698 119 - 1 -CUAGCCACCGCCGCAUCACGUGUGGCACUAUAAAUAUAAUUUACCCGAGGCGCAUGUAAUUGCGGCGCUGCCUAAAUUACCCCGCCUUGAACACACACAUUGUAUAUGUUUAUUAAUAU -...(((((((........)).)))))...(((((((((.......(((((((...((((((..(((...)))..))))))..)))))))...(((.....))).)))))))))...... ( -32.90) >DroSim_CAF1 23914 119 - 1 -CUAGCCACCGCCGCAUCACGUGUGGCACGAUAAAUAUAAUUUACCCGAGGCGCAUGUAAUUGCGGCGCUGCCUAAAUUACCCCGCCUUGAACACACACAUUGUAUAUGUUUAUUAACAU -...(((((((........)).)))))..((((((((((.......(((((((...((((((..(((...)))..))))))..)))))))...(((.....))).))))))))))..... ( -34.90) >DroEre_CAF1 26398 119 - 1 -CUAGCCACCGCCGCAUCACGUGUGGCACGAUAAAUAUAAUUUACCCGAGGCGCAUGUAAUUGCGGCGCUGCCUAAAUUACCCCGCCUUGAACACACACAUUGUAUAUGAUUAUUAACAU -...(((((((........)).)))))..(((((.((((.......(((((((...((((((..(((...)))..))))))..)))))))...(((.....))).)))).)))))..... ( -30.90) >DroYak_CAF1 17355 120 - 1 GCUUGCCACCGCCGCAUCACGUGUGGCACGAUAAAUAUAAUUUACCCGAGGCGCAUGUAAUUGCGGCGCUGCCUAAAUUACCCCGCCUUGAACACACACAUUGUAUAUGUUUAUUAACAU ...((((((((........)).)))))).((((((((((.......(((((((...((((((..(((...)))..))))))..)))))))...(((.....))).))))))))))..... ( -35.30) >consensus _CUAGCCACCGCCGCAUCACGUGUGGCACGAUAAAUAUAAUUUACCCGAGGCGCAUGUAAUUGCGGCGCUGCCUAAAUUACCCCGCCUUGAACACACACAUUGUAUAUGUUUAUUAACAU ..........(((((((...)))))))..((((((((((.......(((((((...((((((..(((...)))..))))))..)))))))...(((.....))).))))))))))..... (-33.16 = -33.56 + 0.40)

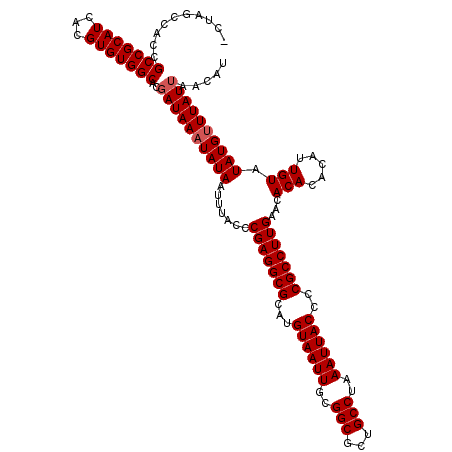
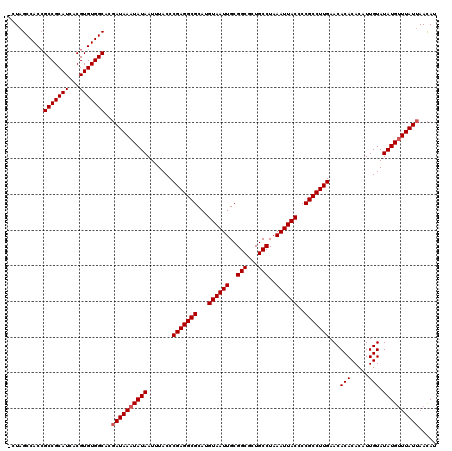
| Location | 2,635,422 – 2,635,539 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.79 |
| Mean single sequence MFE | -37.04 |
| Consensus MFE | -31.64 |
| Energy contribution | -32.24 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.85 |
| SVM decision value | 2.12 |
| SVM RNA-class probability | 0.988461 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2635422 117 - 27905053 GGGUCAUUCCAUAAUUUCGAAAGGACUAGAGGAAACUG---CUAGCCACAGCCGCAUCACGUGUGGCACGAUAAAUAUAAUUUACCCGAGGCGCAUGUAAUUGCGGCGCUGCCUAAAUUA ((((..................((.((((((....)).---))))))...((((((..(((((((.(.((.(((((...)))))..)).).)))))))...))))))...))))...... ( -38.20) >DroSec_CAF1 35738 117 - 1 GGGUCAUUCCAUAAUUUCGAAAGGACUACAGGAAACUG---CUAGCCACCGCCGCAUCACGUGUGGCACUAUAAAUAUAAUUUACCCGAGGCGCAUGUAAUUGCGGCGCUGCCUAAAUUA ((((..................((.((((((....)))---.)))))..(((((((..(((((((.(.(..(((((...)))))...).).)))))))...)))))))..))))...... ( -36.30) >DroSim_CAF1 23954 117 - 1 GGGUCAUUCCAUAAUUUCGAAAGGACUACAGGAAACUG---CUAGCCACCGCCGCAUCACGUGUGGCACGAUAAAUAUAAUUUACCCGAGGCGCAUGUAAUUGCGGCGCUGCCUAAAUUA ((((..................((.((((((....)))---.)))))..(((((((..(((((((.(.((.(((((...)))))..)).).)))))))...)))))))..))))...... ( -39.20) >DroEre_CAF1 26438 116 - 1 GGGUCAUUCCAUAAUUUCGA-AGGACUAGAGGAAACUG---CUAGCCACCGCCGCAUCACGUGUGGCACGAUAAAUAUAAUUUACCCGAGGCGCAUGUAAUUGCGGCGCUGCCUAAAUUA ((((................-.((.((((((....)).---))))))..(((((((..(((((((.(.((.(((((...)))))..)).).)))))))...)))))))..))))...... ( -39.40) >DroYak_CAF1 17395 120 - 1 GGGUCAUUCCAUAAUUUCGAAAGGACUAGAGAAAACUGCUGCUUGCCACCGCCGCAUCACGUGUGGCACGAUAAAUAUAAUUUACCCGAGGCGCAUGUAAUUGCGGCGCUGCCUAAAUUA (((....))).....(((....))).(((........((.....))((.(((((((..(((((((.(.((.(((((...)))))..)).).)))))))...))))))).)).)))..... ( -32.10) >consensus GGGUCAUUCCAUAAUUUCGAAAGGACUAGAGGAAACUG___CUAGCCACCGCCGCAUCACGUGUGGCACGAUAAAUAUAAUUUACCCGAGGCGCAUGUAAUUGCGGCGCUGCCUAAAUUA (((....))).....(((....)))....((....)).....(((.((.(((((((..(((((((.(.((.(((((...)))))..)).).)))))))...))))))).)).)))..... (-31.64 = -32.24 + 0.60)

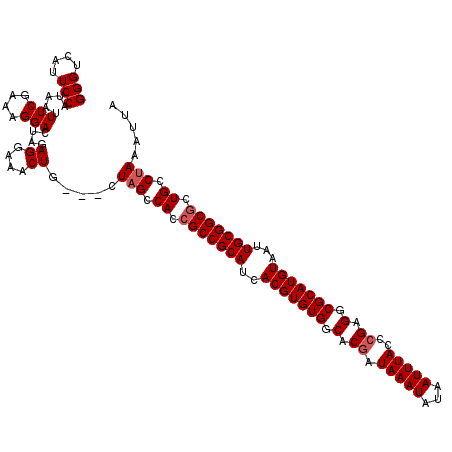
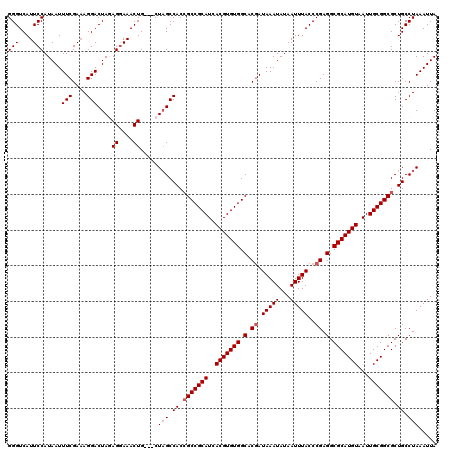
| Location | 2,635,462 – 2,635,579 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.43 |
| Mean single sequence MFE | -36.40 |
| Consensus MFE | -27.80 |
| Energy contribution | -28.20 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.848216 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2635462 117 - 27905053 UUAAUUGGCAGUCCACGGCGGGCUUUGUAAUCACAUUUAUGGGUCAUUCCAUAAUUUCGAAAGGACUAGAGGAAACUG---CUAGCCACAGCCGCAUCACGUGUGGCACGAUAAAUAUAA .........(((((.....)))))(((((.......(((((((....)))))))..(((...((.((((((....)).---))))))...(((((((...))))))).)))....))))) ( -34.20) >DroSec_CAF1 35778 117 - 1 UUAAUUGGCAGGCCACGGCGGGCUUUGUAAUCACAUUUAUGGGUCAUUCCAUAAUUUCGAAAGGACUACAGGAAACUG---CUAGCCACCGCCGCAUCACGUGUGGCACUAUAAAUAUAA .........((((((((((((.........((....(((((((....)))))))....))..((.((((((....)))---.))))).)))))((.....))))))).)).......... ( -37.70) >DroSim_CAF1 23994 117 - 1 UUAAUUGGCAGGCCACGACGGGCUUUGUAAUCACAUUUAUGGGUCAUUCCAUAAUUUCGAAAGGACUACAGGAAACUG---CUAGCCACCGCCGCAUCACGUGUGGCACGAUAAAUAUAA .......(((((((......))).))))........(((((((....)))))))..(((...((.((((((....)))---.)))))...(((((((...))))))).)))......... ( -35.40) >DroEre_CAF1 26478 116 - 1 UUAAUUGGCAGGCCACAGCGAGCUUUGUAAUCACAUUUAUGGGUCAUUCCAUAAUUUCGA-AGGACUAGAGGAAACUG---CUAGCCACCGCCGCAUCACGUGUGGCACGAUAAAUAUAA ...((((....(((((((((.((.......((....(((((((....)))))))....))-.((.((((((....)).---))))))...)))))......)))))).))))........ ( -34.20) >DroYak_CAF1 17435 120 - 1 UUAAUUGGCAGGCCACGGCGGGCUUUGUAAUCACAUUUAUGGGUCAUUCCAUAAUUUCGAAAGGACUAGAGAAAACUGCUGCUUGCCACCGCCGCAUCACGUGUGGCACGAUAAAUAUAA .....((((((((...(((((.(((((.........(((((((....))))))).(((....))).)))))....)))))))))))))..(((((((...)))))))............. ( -40.50) >consensus UUAAUUGGCAGGCCACGGCGGGCUUUGUAAUCACAUUUAUGGGUCAUUCCAUAAUUUCGAAAGGACUAGAGGAAACUG___CUAGCCACCGCCGCAUCACGUGUGGCACGAUAAAUAUAA .....(((((((((......)))))...........(((((((....))))))).(((....)))....((....)).......))))..(((((((...)))))))............. (-27.80 = -28.20 + 0.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:50:56 2006