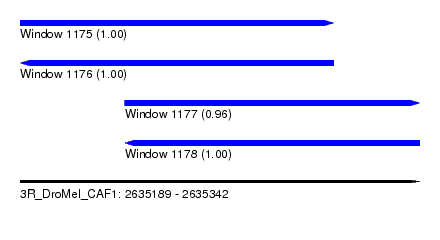
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 2,635,189 – 2,635,342 |
| Length | 153 |
| Max. P | 0.999908 |

| Location | 2,635,189 – 2,635,309 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.17 |
| Mean single sequence MFE | -32.15 |
| Consensus MFE | -28.61 |
| Energy contribution | -29.41 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.99 |
| Structure conservation index | 0.89 |
| SVM decision value | 3.60 |
| SVM RNA-class probability | 0.999436 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2635189 120 + 27905053 CGCUUGCCAUCAGCAACACUUUUUCGAUGGAUAUUGAACCACACAAACCUCUCUAGAACCGAAUUUGUCCUUAAAGCUAGUAUAAAACAAAUUCGGAUCUACAGGGUAGACGUGGAGCAA .(((..(((((..............)))))........((((....(((.((.((((.((((((((((..................)))))))))).)))).)))))....))))))).. ( -33.31) >DroSec_CAF1 35507 120 + 1 CGCUUGCCAUCAGCUACACUUUUUCGAUGGAUAUUGAACCACACAAACCUCUCUAGAACCGAAUUUGUCCUUAAAGCUAGUAUAAAACAAAUUCGGAUCUACAGGGUAGACGUGGAGCAA .(((..(((((..............)))))........((((....(((.((.((((.((((((((((..................)))))))))).)))).)))))....))))))).. ( -33.31) >DroSim_CAF1 23724 120 + 1 CGCUUGCCAUAAGCUACACUUUUUCGAUGGAUAUUGAACCACACAAACCUCUCUAGAACCGAAUUUGUCCUUAAAGCUAGUAUAAAACAAAUUCGGAUCUACAGGGUAGACGUGGAGCAA ............(((.......((((((....))))))((((....(((.((.((((.((((((((((..................)))))))))).)))).)))))....))))))).. ( -30.87) >DroEre_CAF1 26210 120 + 1 CGCUUGACAUCAGCUUCACUUUUCCGAUGGAUAUUGAACCACACAAACCUCUCUAGAACCGAAUUUGUCCUUAAAGCUAGUAUAAAACAAAUUCGGAUCUACAGGGUAGACGUAGAGCAA .((((.((.((.(.((((....(((...)))...)))).)......(((.((.((((.((((((((((..................)))))))))).)))).))))).)).)).)))).. ( -30.57) >DroYak_CAF1 17160 120 + 1 CGCUUGACAUCAGCUUCACUUUUCCGAUGGAUAUUGAACCACACAAACCUCUCUAGAACCGAAUUUGUCCUUAAAACUAGUAUAAAACAAAUUCGGAUCUACAGGGUAGACGUGGAGCAA .((((.((.((.(.((((....(((...)))...)))).)......(((.((.((((.((((((((((..................)))))))))).)))).))))).)).)).)))).. ( -32.67) >consensus CGCUUGCCAUCAGCUACACUUUUUCGAUGGAUAUUGAACCACACAAACCUCUCUAGAACCGAAUUUGUCCUUAAAGCUAGUAUAAAACAAAUUCGGAUCUACAGGGUAGACGUGGAGCAA .(((..(((((..............)))))........((((....(((.((.((((.((((((((((..................)))))))))).)))).)))))....))))))).. (-28.61 = -29.41 + 0.80)

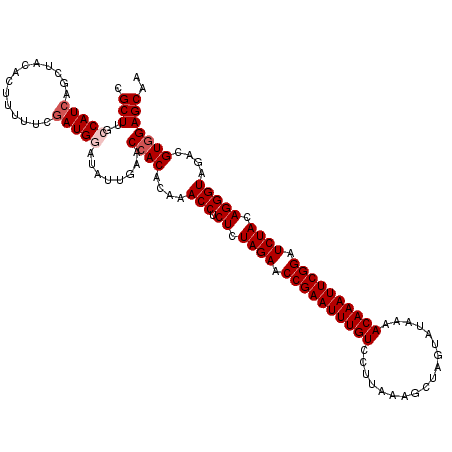
| Location | 2,635,189 – 2,635,309 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.17 |
| Mean single sequence MFE | -38.38 |
| Consensus MFE | -36.14 |
| Energy contribution | -35.98 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.00 |
| Structure conservation index | 0.94 |
| SVM decision value | 4.49 |
| SVM RNA-class probability | 0.999908 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2635189 120 - 27905053 UUGCUCCACGUCUACCCUGUAGAUCCGAAUUUGUUUUAUACUAGCUUUAAGGACAAAUUCGGUUCUAGAGAGGUUUGUGUGGUUCAAUAUCCAUCGAAAAAGUGUUGCUGAUGGCAAGCG ..((((((((...(((((.((((.(((((((((((((............))))))))))))).)))).)).)))...)))))........((((((..((....))..))))))..))). ( -39.00) >DroSec_CAF1 35507 120 - 1 UUGCUCCACGUCUACCCUGUAGAUCCGAAUUUGUUUUAUACUAGCUUUAAGGACAAAUUCGGUUCUAGAGAGGUUUGUGUGGUUCAAUAUCCAUCGAAAAAGUGUAGCUGAUGGCAAGCG ..((((((((...(((((.((((.(((((((((((((............))))))))))))).)))).)).)))...)))))........((((((............))))))..))). ( -38.80) >DroSim_CAF1 23724 120 - 1 UUGCUCCACGUCUACCCUGUAGAUCCGAAUUUGUUUUAUACUAGCUUUAAGGACAAAUUCGGUUCUAGAGAGGUUUGUGUGGUUCAAUAUCCAUCGAAAAAGUGUAGCUUAUGGCAAGCG (((..(((((...(((((.((((.(((((((((((((............))))))))))))).)))).)).)))...)))))..)))...................((((.....)))). ( -36.80) >DroEre_CAF1 26210 120 - 1 UUGCUCUACGUCUACCCUGUAGAUCCGAAUUUGUUUUAUACUAGCUUUAAGGACAAAUUCGGUUCUAGAGAGGUUUGUGUGGUUCAAUAUCCAUCGGAAAAGUGAAGCUGAUGUCAAGCG ..(((..(((((.(((((.((((.(((((((((((((............))))))))))))).)))).)).)))......(.((((...(((...)))....)))).).)))))..))). ( -38.30) >DroYak_CAF1 17160 120 - 1 UUGCUCCACGUCUACCCUGUAGAUCCGAAUUUGUUUUAUACUAGUUUUAAGGACAAAUUCGGUUCUAGAGAGGUUUGUGUGGUUCAAUAUCCAUCGGAAAAGUGAAGCUGAUGUCAAGCG ..((((((((...(((((.((((.(((((((((((((............))))))))))))).)))).)).)))...))))).........((((((..........))))))...))). ( -39.00) >consensus UUGCUCCACGUCUACCCUGUAGAUCCGAAUUUGUUUUAUACUAGCUUUAAGGACAAAUUCGGUUCUAGAGAGGUUUGUGUGGUUCAAUAUCCAUCGAAAAAGUGUAGCUGAUGGCAAGCG (((..(((((...(((((.((((.(((((((((((((............))))))))))))).)))).)).)))...)))))..)))...................(((.......))). (-36.14 = -35.98 + -0.16)

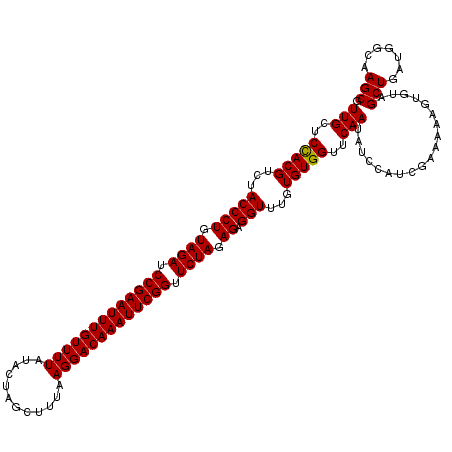
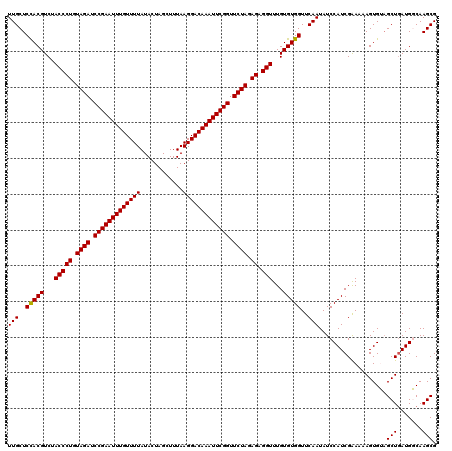
| Location | 2,635,229 – 2,635,342 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.32 |
| Mean single sequence MFE | -29.49 |
| Consensus MFE | -25.15 |
| Energy contribution | -25.59 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.963511 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2635229 113 + 27905053 ACACAAACCUCUCUAGAACCGAAUUUGUCCUUAAAGCUAGUAUAAAACAAAUUCGGAUCUACAGGGUAGACGUGGAGCAAACUGAUGAGGAACA-------GUCGCUGGGGCGAAAUGAA .......((((..(((..((((((((((..................)))))))))).(((((.(......)))))).....)))..))))....-------.((((....))))...... ( -28.17) >DroSec_CAF1 35547 113 + 1 ACACAAACCUCUCUAGAACCGAAUUUGUCCUUAAAGCUAGUAUAAAACAAAUUCGGAUCUACAGGGUAGACGUGGAGCAAACUGAUGAGGAACA-------GUCGCUGGGGCGAAAUGAA .......((((..(((..((((((((((..................)))))))))).(((((.(......)))))).....)))..))))....-------.((((....))))...... ( -28.17) >DroSim_CAF1 23764 113 + 1 ACACAAACCUCUCUAGAACCGAAUUUGUCCUUAAAGCUAGUAUAAAACAAAUUCGGAUCUACAGGGUAGACGUGGAGCAAACUGAUGAGGAACA-------GUCGCUGGGGCGAAAUGAA .......((((..(((..((((((((((..................)))))))))).(((((.(......)))))).....)))..))))....-------.((((....))))...... ( -28.17) >DroEre_CAF1 26250 112 + 1 ACACAAACCUCUCUAGAACCGAAUUUGUCCUUAAAGCUAGUAUAAAACAAAUUCGGAUCUACAGGGUAGACGUAGAGCAAACUGAUGAGGAACGU--------CGGAGGGGCGAAAUGAA .......((((((((((.((((((((((..................)))))))))).)))........(((((....((......))....))))--------))))))))......... ( -29.57) >DroYak_CAF1 17200 120 + 1 ACACAAACCUCUCUAGAACCGAAUUUGUCCUUAAAACUAGUAUAAAACAAAUUCGGAUCUACAGGGUAGACGUGGAGCAAAGUGAUGAGGAACGUCAGUUCGUCGCAGGGGCGAAAUGAA ......(((.((.((((.((((((((((..................)))))))))).)))).))))).(((((....((......))....)))))..((((((.....))))))..... ( -33.37) >consensus ACACAAACCUCUCUAGAACCGAAUUUGUCCUUAAAGCUAGUAUAAAACAAAUUCGGAUCUACAGGGUAGACGUGGAGCAAACUGAUGAGGAACA_______GUCGCUGGGGCGAAAUGAA .......((((..(((..((((((((((..................)))))))))).(((((.(......)))))).....)))..))))............((((....))))...... (-25.15 = -25.59 + 0.44)

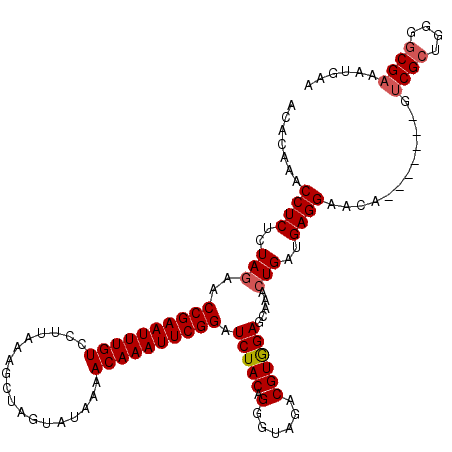
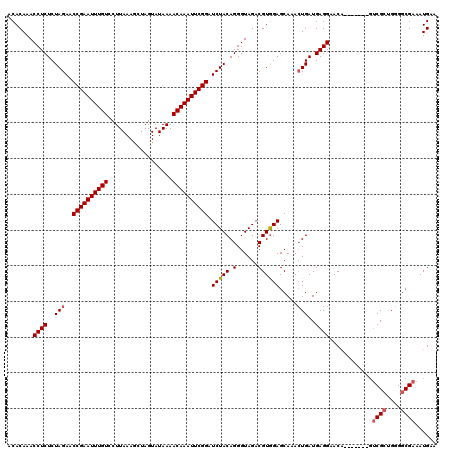
| Location | 2,635,229 – 2,635,342 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.32 |
| Mean single sequence MFE | -34.44 |
| Consensus MFE | -30.26 |
| Energy contribution | -30.50 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.55 |
| Structure conservation index | 0.88 |
| SVM decision value | 4.36 |
| SVM RNA-class probability | 0.999880 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2635229 113 - 27905053 UUCAUUUCGCCCCAGCGAC-------UGUUCCUCAUCAGUUUGCUCCACGUCUACCCUGUAGAUCCGAAUUUGUUUUAUACUAGCUUUAAGGACAAAUUCGGUUCUAGAGAGGUUUGUGU .............((((((-------((........))).))))).((((...(((((.((((.(((((((((((((............))))))))))))).)))).)).))).)))). ( -35.20) >DroSec_CAF1 35547 113 - 1 UUCAUUUCGCCCCAGCGAC-------UGUUCCUCAUCAGUUUGCUCCACGUCUACCCUGUAGAUCCGAAUUUGUUUUAUACUAGCUUUAAGGACAAAUUCGGUUCUAGAGAGGUUUGUGU .............((((((-------((........))).))))).((((...(((((.((((.(((((((((((((............))))))))))))).)))).)).))).)))). ( -35.20) >DroSim_CAF1 23764 113 - 1 UUCAUUUCGCCCCAGCGAC-------UGUUCCUCAUCAGUUUGCUCCACGUCUACCCUGUAGAUCCGAAUUUGUUUUAUACUAGCUUUAAGGACAAAUUCGGUUCUAGAGAGGUUUGUGU .............((((((-------((........))).))))).((((...(((((.((((.(((((((((((((............))))))))))))).)))).)).))).)))). ( -35.20) >DroEre_CAF1 26250 112 - 1 UUCAUUUCGCCCCUCCG--------ACGUUCCUCAUCAGUUUGCUCUACGUCUACCCUGUAGAUCCGAAUUUGUUUUAUACUAGCUUUAAGGACAAAUUCGGUUCUAGAGAGGUUUGUGU .......(((......(--------((((....((......))....))))).(((((.((((.(((((((((((((............))))))))))))).)))).)).)))..))). ( -31.50) >DroYak_CAF1 17200 120 - 1 UUCAUUUCGCCCCUGCGACGAACUGACGUUCCUCAUCACUUUGCUCCACGUCUACCCUGUAGAUCCGAAUUUGUUUUAUACUAGUUUUAAGGACAAAUUCGGUUCUAGAGAGGUUUGUGU .......(((....)))((((((((((((....((......))....)))))....((.((((.(((((((((((((............))))))))))))).)))).)).))))))).. ( -35.10) >consensus UUCAUUUCGCCCCAGCGAC_______UGUUCCUCAUCAGUUUGCUCCACGUCUACCCUGUAGAUCCGAAUUUGUUUUAUACUAGCUUUAAGGACAAAUUCGGUUCUAGAGAGGUUUGUGU ......((((....))))............................((((...(((((.((((.(((((((((((((............))))))))))))).)))).)).))).)))). (-30.26 = -30.50 + 0.24)

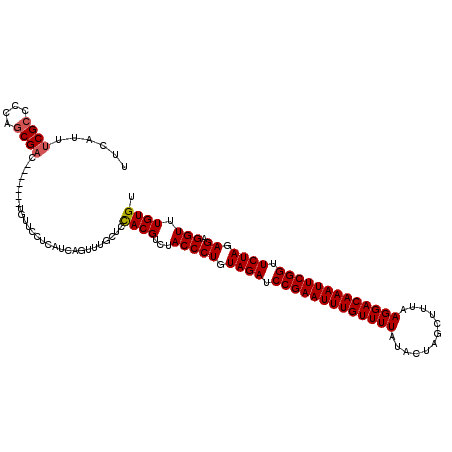
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:50:52 2006