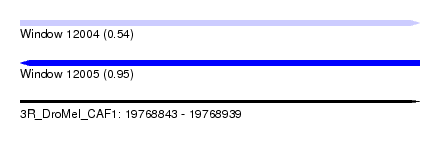
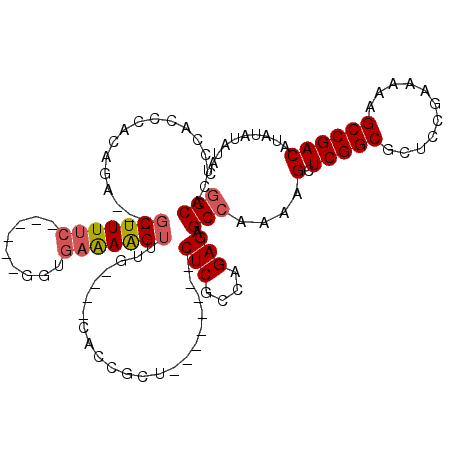
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 19,768,843 – 19,768,939 |
| Length | 96 |
| Max. P | 0.947013 |

| Location | 19,768,843 – 19,768,939 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 80.15 |
| Mean single sequence MFE | -39.84 |
| Consensus MFE | -16.71 |
| Energy contribution | -17.55 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.42 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.539563 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19768843 96 + 27905053 GUAUAUAUAUGUCGGCUUUUUCGGAGCGCCGAGCUUUUUGCUCUCCGGCGAG--------AGCGGUG----CAAACUUUUCACC------GAAAAGC---UCUGUGGGUGGAGUGCC .............(((...(((.(..(((.(((((((((((((((....)))--------)))((((----.((....))))))------)))))))---)).)))..).))).))) ( -35.10) >DroSec_CAF1 8850 96 + 1 GUAUAUAUAUGUCGGCUUUUUCGGAGAGCCGAGCUUUUGGCUCUCUGGCGAG--------AGCGGUG----CAAACUUUUCACC------GAAAAGC---UCUGUGGGUGGAGUGCC ((((.....((((.((((((((((((((((((....)))))))))))).)))--------))).).)----)).....((((((------.(.....---....).)))))))))). ( -34.70) >DroSim_CAF1 9180 96 + 1 GUAUAUAUAUGUCGGCUUUUUCGGAGAGCCGAGCUUUUGGCUCUCUGGCGAG--------AGCGAUG----CAAACUUUUCACC------GAAAAGC---UCUGUGGGUGGAGUGCC ..........((((.(((((((((((((((((....)))))))))))).)))--------))))))(----((..((.(((((.------((.....---)).))))).))..))). ( -34.80) >DroEre_CAF1 8866 113 + 1 GCAUAUAUAUGUCGGCUUUUUCGGACCGCCGAGCUUUUGGCUCUCUCGCGAGUGCGCUGGAGAG----GAGUACACCUCUCACCGAGAGCGAAAAGCUCAUCUGCGGGUGGGGUGCC ((((....)))).(((..(..((..((((.((((((((.((((((..(((....)))..(((((----(......))))))...)))))).))))))))....)))).))..).))) ( -50.00) >DroYak_CAF1 9030 108 + 1 GCAUAUAUAUGUCGGCUUUUUCGGAGCGCCGAGCUUUCGGCUCUCUCGCGAGU-CGCGAGAGCGGUGAGAGCACUCUUCUCUC----AGCU-AAAGC---UCUGCGGGUGGAGUGCC ((.........((((((((....))).)))))(((((((.(.((((((((...-)))))))).).)))))))(((((.(((.(----((..-.....---.))).))).))))))). ( -44.60) >consensus GUAUAUAUAUGUCGGCUUUUUCGGAGCGCCGAGCUUUUGGCUCUCUGGCGAG________AGCGGUG____CAAACUUUUCACC______GAAAAGC___UCUGUGGGUGGAGUGCC .............(((....((((((.(((((....))))).)))))).............................(((((((......................))))))).))) (-16.71 = -17.55 + 0.84)

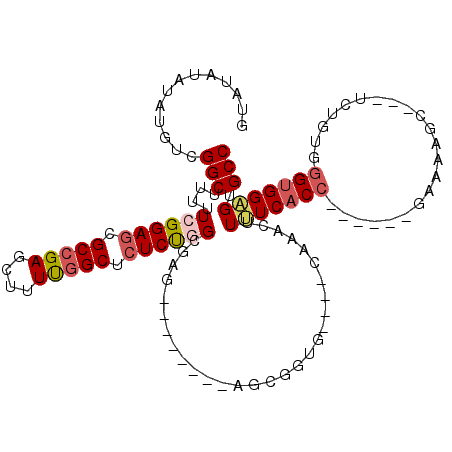

| Location | 19,768,843 – 19,768,939 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 80.15 |
| Mean single sequence MFE | -34.98 |
| Consensus MFE | -16.84 |
| Energy contribution | -17.24 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.48 |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.947013 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19768843 96 - 27905053 GGCACUCCACCCACAGA---GCUUUUC------GGUGAAAAGUUUG----CACCGCU--------CUCGCCGGAGAGCAAAAAGCUCGGCGCUCCGAAAAAGCCGACAUAUAUAUAC (((............((---((((((.------((((.........----))))(((--------(((....)))))).))))))))((....))......)))............. ( -30.70) >DroSec_CAF1 8850 96 - 1 GGCACUCCACCCACAGA---GCUUUUC------GGUGAAAAGUUUG----CACCGCU--------CUCGCCAGAGAGCCAAAAGCUCGGCUCUCCGAAAAAGCCGACAUAUAUAUAC (((............((---((((((.------((((.........----))))(((--------(((....)))))).))))))))((....))......)))............. ( -30.40) >DroSim_CAF1 9180 96 - 1 GGCACUCCACCCACAGA---GCUUUUC------GGUGAAAAGUUUG----CAUCGCU--------CUCGCCAGAGAGCCAAAAGCUCGGCUCUCCGAAAAAGCCGACAUAUAUAUAC (((.(((........))---).(((((------(((((..(((...----....)))--------.))))).(((((((........))))))).))))).)))............. ( -28.60) >DroEre_CAF1 8866 113 - 1 GGCACCCCACCCGCAGAUGAGCUUUUCGCUCUCGGUGAGAGGUGUACUC----CUCUCCAGCGCACUCGCGAGAGAGCCAAAAGCUCGGCGGUCCGAAAAAGCCGACAUAUAUAUGC (((.......((((....((((((((.((((((.(.((((((......)----))))))..(((....))).)))))).)))))))).)))).........)))............. ( -45.19) >DroYak_CAF1 9030 108 - 1 GGCACUCCACCCGCAGA---GCUUU-AGCU----GAGAGAAGAGUGCUCUCACCGCUCUCGCG-ACUCGCGAGAGAGCCGAAAGCUCGGCGCUCCGAAAAAGCCGACAUAUAUAUGC (((..((....(((.((---(((((-.(((----(((((.......)))))....((((((((-...)))))))))))..))))))).)))....))....)))............. ( -40.00) >consensus GGCACUCCACCCACAGA___GCUUUUC______GGUGAAAAGUUUG____CACCGCU________CUCGCCAGAGAGCCAAAAGCUCGGCGCUCCGAAAAAGCCGACAUAUAUAUAC (((.................(((((((.........)))))))......................(((....))).)))....(.(((((...........)))))).......... (-16.84 = -17.24 + 0.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:45:41 2006