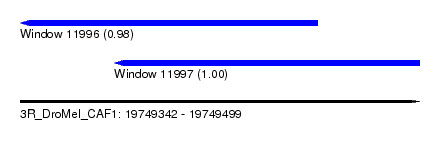
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 19,749,342 – 19,749,499 |
| Length | 157 |
| Max. P | 0.996290 |

| Location | 19,749,342 – 19,749,459 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.55 |
| Mean single sequence MFE | -36.70 |
| Consensus MFE | -28.78 |
| Energy contribution | -29.22 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.80 |
| SVM RNA-class probability | 0.977845 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19749342 117 - 27905053 CCCAGUUCUUCAGCGGAUGAUCCUGCUGCAGGACAGUCAUGCAGCGAAAGUUGUACGGUUGGAAUACCAACAUGAAGUAGUAUCCCUACGGAAGAAGCUUUUAUCAAUUGAAGA---AAA .((.(((((.((((((......)))))).)))))..(((((((((....)))))...(((((....))))))))).((((.....))))))......((((........)))).---... ( -35.90) >DroSec_CAF1 16602 118 - 1 CCCAGUUCUUCAGCGGAUGAUCCUGCUGCAGGACGGUCAUGCAGCGAAAGUUGUACGGUUGGAAUACCAGCAUGAAGUAGUAUCCCUACGAAAGAAGCUUUUAUCAAUUGAAAA--AAAA .((.(((((.((((((......)))))).)))))))(((((((((....))))))..(((((....))))).(((.((((.....))))(((((...))))).)))..)))...--.... ( -34.60) >DroSim_CAF1 19408 120 - 1 CCCAGUUCUUCAGCGGAUGAUCCUGCUGCAGGACGGUCAUGCAGCGAAAGUUGUACGGUUGGAAUACCAACAUGAAGUAGUAUCCCUACGAAAGAAGCUUUUAUCAAUUGAAAAAAAAAA .((.(((((.((((((......)))))).)))))))(((((((((....))))))..(((((....))))).(((.((((.....))))(((((...))))).)))..)))......... ( -34.60) >DroEre_CAF1 29700 117 - 1 CCCAGUUCUUCAGCGGAUGAUCCUGCUGCAGGACGGUCAUGCAGCGAAAGUUGUACGGUUGGAAUGCCAACAUGAAGUAGUAUCCCUGAGGGAGAAGGUUUCCUCAAUUGGAAA---UAU .((.(((((.((((((......)))))).)))))))(((((((((....)))))...(((((....))))))))).......(((.((((((((....))))))))...)))..---... ( -42.50) >DroYak_CAF1 30143 117 - 1 CCCAGUUCUUAAGAGGAUGAUCCUGCUGCAAAACGGUCAUGCAGCGAAAGUUGUACGGUUGGAAUGCCAACAUGAAGUAGUAUCCCUGAGGACGAAGGUUUUCUCAAUAGGAAA---UAU .....((((((((.(((((...(((((.((.........((((((....))))))..(((((....))))).)).))))))))))))((((((....))))))....)))))).---... ( -35.90) >consensus CCCAGUUCUUCAGCGGAUGAUCCUGCUGCAGGACGGUCAUGCAGCGAAAGUUGUACGGUUGGAAUACCAACAUGAAGUAGUAUCCCUACGGAAGAAGCUUUUAUCAAUUGAAAA___AAA ....(((((.((((((......)))))).)))))..(((((((((....)))))...(((((....))))))))).((((.....))))............................... (-28.78 = -29.22 + 0.44)

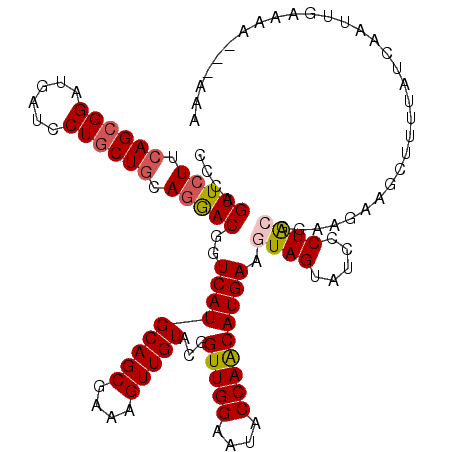
| Location | 19,749,379 – 19,749,499 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.17 |
| Mean single sequence MFE | -40.56 |
| Consensus MFE | -38.20 |
| Energy contribution | -38.52 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.68 |
| SVM RNA-class probability | 0.996290 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19749379 120 - 27905053 ACGAUCUUGUUUAUGUAGUAAGCAUAGCUGAUGCAGCGCUCCCAGUUCUUCAGCGGAUGAUCCUGCUGCAGGACAGUCAUGCAGCGAAAGUUGUACGGUUGGAAUACCAACAUGAAGUAG .......(((((((...)))))))..((((...))))(((....(((((.((((((......)))))).)))))..(((((((((....)))))...(((((....)))))))))))).. ( -37.60) >DroSec_CAF1 16640 120 - 1 ACGAUCUUGUUUAUGUAGUAGGCAUAGCUGAUGCAGCGCUCCCAGUUCUUCAGCGGAUGAUCCUGCUGCAGGACGGUCAUGCAGCGAAAGUUGUACGGUUGGAAUACCAGCAUGAAGUAG ..((((..((((.((((((((((((.(((((.(.(((.......)))).)))))..)))..))))))))))))))))).((((((....))))))..(((((....)))))......... ( -42.30) >DroSim_CAF1 19448 120 - 1 ACGAUCUUGUUUAUGUAGUAGGCAUAGCUGAUGCAGCGCUCCCAGUUCUUCAGCGGAUGAUCCUGCUGCAGGACGGUCAUGCAGCGAAAGUUGUACGGUUGGAAUACCAACAUGAAGUAG ..((((..((((.((((((((((((.(((((.(.(((.......)))).)))))..)))..))))))))))))))))).((((((....))))))..(((((....)))))......... ( -42.30) >DroEre_CAF1 29737 120 - 1 ACGAUCUUGUUUAUGUAGUAGGCAUAGCUGAUGCAGCGCUCCCAGUUCUUCAGCGGAUGAUCCUGCUGCAGGACGGUCAUGCAGCGAAAGUUGUACGGUUGGAAUGCCAACAUGAAGUAG ..((((..((((.((((((((((((.(((((.(.(((.......)))).)))))..)))..))))))))))))))))).((((((....))))))..(((((....)))))......... ( -42.50) >DroYak_CAF1 30180 120 - 1 ACGAUCUUGUUUAUGUAGUAGGCAUAGCUGAUGCAGCGCUCCCAGUUCUUAAGAGGAUGAUCCUGCUGCAAAACGGUCAUGCAGCGAAAGUUGUACGGUUGGAAUGCCAACAUGAAGUAG ..((((..((((.(((((((((....((((...)))).......(((((....)))))...))))))))))))))))).((((((....))))))..(((((....)))))......... ( -38.10) >consensus ACGAUCUUGUUUAUGUAGUAGGCAUAGCUGAUGCAGCGCUCCCAGUUCUUCAGCGGAUGAUCCUGCUGCAGGACGGUCAUGCAGCGAAAGUUGUACGGUUGGAAUACCAACAUGAAGUAG ..((((..((((.((((((((((((.(((((.(.(((.......)))).)))))..)))..))))))))))))))))).((((((....))))))..(((((....)))))......... (-38.20 = -38.52 + 0.32)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:45:34 2006