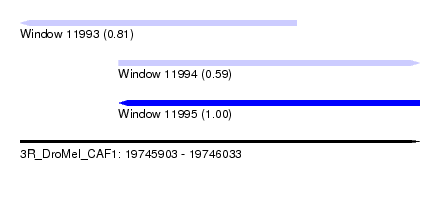
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 19,745,903 – 19,746,033 |
| Length | 130 |
| Max. P | 0.996010 |

| Location | 19,745,903 – 19,745,993 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 81.79 |
| Mean single sequence MFE | -32.67 |
| Consensus MFE | -20.65 |
| Energy contribution | -20.43 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.805336 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19745903 90 - 27905053 CAAUAAAGUCAGCGGCAACAACGGGAGUAACGGCAGCAACUGUUGCUGCUGCAA---CUGC---UGCUACUGGGGCAACGCAUUGCAUGUCGAC------AA .......(((.(((((((...(((.(((..((((((((.....))))))))..)---)).)---))....((.(....).)))))).))).)))------.. ( -32.10) >DroSec_CAF1 13130 90 - 1 CAAUAAAGUCAGUGGCAACAACGGGAGUAACGGCGGCAACUGUUGCUGCUGCAA---CGGC---UGCUACUGGGGCAACGCAUUGCAUGUCGAC------AA .......(((.(..(((.((((((..((.......))..)))))).)))..)..---.(((---(((...((.(....).))..))).))))))------.. ( -31.40) >DroSim_CAF1 15268 90 - 1 CAAUAAAGUCAGCGGCAACAACGGGAGUAACGGCGGCAACUGUUGCUGCUGCAA---CGGC---UGCUACUGGGGCAACGCAUUGCAUGUCGAC------AA .......(((.((((((.((((((..((.......))..)))))).))))))..---.(((---(((...((.(....).))..))).))))))------.. ( -33.90) >DroEre_CAF1 26202 90 - 1 CAAUAAAGUCAGCGGCAACAACGGGAGCAACGGCAGCAACUGUUGCUGCUGCAA---CUGC---UGCUACUGGGGCAACGCAUUGCAUGUCGAC------AA .......((((((((((....(((.((((((((......)))))))).)))...---.)))---))))..((.(....).)).........)))------.. ( -33.60) >DroYak_CAF1 26378 99 - 1 CAAUAAAGUCAGCGGCAACAACGGGAGUAACGGCAGCAACUGUUGCUGCUGCAA---CUGCUACUGCUACUGGGGCAACGCAUUGCAUGUCCACAACAACAA .........(((..(((.....((.(((..((((((((.....))))))))..)---)).))..)))..)))(((((..((...)).))))).......... ( -30.20) >DroAna_CAF1 26488 81 - 1 CAGUUAAAUCGACGG---------------CGGCGGCAACUGUUGCUGCUGCUGUCUCCGUCACUGUUGCUAGUGCGGCGCAUUGCCUGCCGAC------AA ..........(((((---------------((((((((.....)))))))))))))...(((...(((((....)))))(((.....))).)))------.. ( -34.80) >consensus CAAUAAAGUCAGCGGCAACAACGGGAGUAACGGCAGCAACUGUUGCUGCUGCAA___CUGC___UGCUACUGGGGCAACGCAUUGCAUGUCGAC______AA ............(((((..............(((((((.....)))))))((((.....((...((((.....))))..)).)))).))))).......... (-20.65 = -20.43 + -0.22)

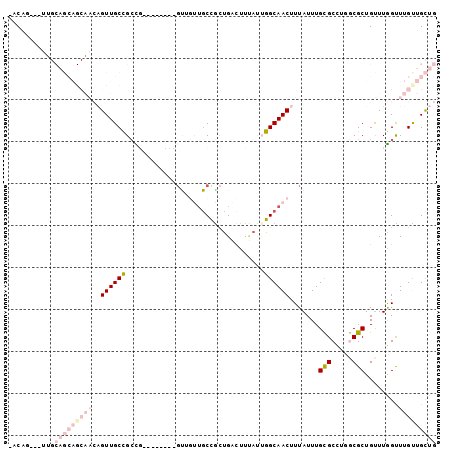
| Location | 19,745,935 – 19,746,033 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 68.36 |
| Mean single sequence MFE | -28.62 |
| Consensus MFE | -12.48 |
| Energy contribution | -14.93 |
| Covariance contribution | 2.45 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.44 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.594432 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19745935 98 + 27905053 -GCAG---UUGCAGCAGCAACAGUUGCUGCCGUUACUCCCGUUGUUGCCGCUGACUUUAUUGGCAACUUUAUUUGCGCCUGGCGCUGUUUGGUUUGUUGCUG -.(((---(.((((((((...(((.((....)).)))...)))))))).))))........((((((.(((...((((...))))....)))...)))))). ( -31.00) >DroVir_CAF1 26525 74 + 1 -ACA------------------GUUGCUGCU---------GCGGCUGCUGCUGACUUUAAAGGCAACUUUAUUUGUGCAGCACGCUGUUUGGUUUGCUGUUG -(((------------------((.(((...---------(((((((((((......(((((....))))).....)))))).)))))..)))..))))).. ( -30.30) >DroSec_CAF1 13162 98 + 1 -GCCG---UUGCAGCAGCAACAGUUGCCGCCGUUACUCCCGUUGUUGCCACUGACUUUAUUGGCAACUUUAUUUGCGCCUGGCGCUGUUUGGUUUGUUGCUG -....---...(((((((((((((.((((.(((..........(((((((.((....)).))))))).......))).).))))))))......)))))))) ( -30.13) >DroYak_CAF1 26418 99 + 1 AGCAG---UUGCAGCAGCAACAGUUGCUGCCGUUACUCCCGUUGUUGCCGCUGACUUUAUUGGCAACUUUAUUUGCGCCUGGCGCUGUUUGGUUUGUUGCUG (((((---..((((((((....))))))))........(((..(((((((.((....)).))))))).......((((...))))....)))....))))). ( -31.50) >DroMoj_CAF1 26812 74 + 1 -ACA------------------GUUGCCGAU---------GUGUUUGCUGCUGGUUUCACAGGCAACUUUAUUUGUGCUGUACGCUGCUUAGCUUGUUGCUG -..(------------------((((((..(---------(((...((.....))..)))))))))))........((..((.(((....))).))..)).. ( -18.10) >DroAna_CAF1 26522 87 + 1 GACGGAGACAGCAGCAGCAACAGUUGCCGCCG---------------CCGUCGAUUUAACUGGCAACUUUAUUUGCGCCUGGCGCUGUUUGGCUUGCUGCUG ........((((((((((((.((((((((...---------------.............))))))))....))))(((..((...))..))).)))))))) ( -30.69) >consensus _ACAG___UUGCAGCAGCAACAGUUGCCGCCG________GUUGUUGCCGCUGACUUUAUUGGCAACUUUAUUUGCGCCUGGCGCUGUUUGGUUUGUUGCUG ...........(((((((((..((((((.................................)))))).......(((.....)))........))))))))) (-12.48 = -14.93 + 2.45)

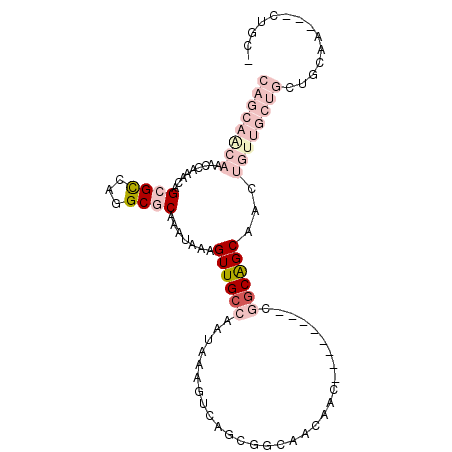
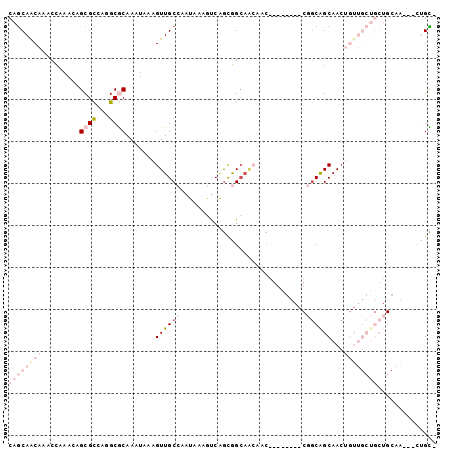
| Location | 19,745,935 – 19,746,033 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 68.36 |
| Mean single sequence MFE | -27.36 |
| Consensus MFE | -10.63 |
| Energy contribution | -13.58 |
| Covariance contribution | 2.94 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.39 |
| SVM decision value | 2.64 |
| SVM RNA-class probability | 0.996010 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19745935 98 - 27905053 CAGCAACAAACCAAACAGCGCCAGGCGCAAAUAAAGUUGCCAAUAAAGUCAGCGGCAACAACGGGAGUAACGGCAGCAACUGUUGCUGCUGCAA---CUGC- ..((......((.....((((...)))).......((((((............))))))...)).(((..((((((((.....))))))))..)---))))- ( -31.50) >DroVir_CAF1 26525 74 - 1 CAACAGCAAACCAAACAGCGUGCUGCACAAAUAAAGUUGCCUUUAAAGUCAGCAGCAGCCGC---------AGCAGCAAC------------------UGU- ..............((((..((((((.........((((.((....)).)))).((....))---------.)))))).)------------------)))- ( -18.90) >DroSec_CAF1 13162 98 - 1 CAGCAACAAACCAAACAGCGCCAGGCGCAAAUAAAGUUGCCAAUAAAGUCAGUGGCAACAACGGGAGUAACGGCGGCAACUGUUGCUGCUGCAA---CGGC- ((((......((.....((((...)))).......(((((((..........)))))))...)).((((((((......))))))))))))...---....- ( -31.20) >DroYak_CAF1 26418 99 - 1 CAGCAACAAACCAAACAGCGCCAGGCGCAAAUAAAGUUGCCAAUAAAGUCAGCGGCAACAACGGGAGUAACGGCAGCAACUGUUGCUGCUGCAA---CUGCU .(((......((.....((((...)))).......((((((............))))))...)).(((..((((((((.....))))))))..)---))))) ( -33.20) >DroMoj_CAF1 26812 74 - 1 CAGCAACAAGCUAAGCAGCGUACAGCACAAAUAAAGUUGCCUGUGAAACCAGCAGCAAACAC---------AUCGGCAAC------------------UGU- ..((.....(((....))).....))........(((((((((((..............)))---------)..))))))------------------)..- ( -15.94) >DroAna_CAF1 26522 87 - 1 CAGCAGCAAGCCAAACAGCGCCAGGCGCAAAUAAAGUUGCCAGUUAAAUCGACGG---------------CGGCGGCAACUGUUGCUGCUGCUGUCUCCGUC ((((((((((((.....((((...)))).......((((((.(((.....)))))---------------))))))).....)))))))))........... ( -33.40) >consensus CAGCAACAAACCAAACAGCGCCAGGCGCAAAUAAAGUUGCCAAUAAAGUCAGCGGCAACAAC________CGGCAGCAACUGUUGCUGCUGCAA___CUGC_ ((((((((.........((((...)))).......((((((..............................))))))...)))))))).............. (-10.63 = -13.58 + 2.94)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:45:32 2006