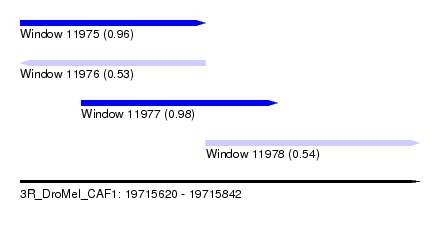
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 19,715,620 – 19,715,842 |
| Length | 222 |
| Max. P | 0.983766 |

| Location | 19,715,620 – 19,715,723 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 92.53 |
| Mean single sequence MFE | -31.04 |
| Consensus MFE | -25.39 |
| Energy contribution | -26.19 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.56 |
| SVM RNA-class probability | 0.963874 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19715620 103 + 27905053 UUCGCGGGCUUCUCUU-AAAGUGAAGGCAGUGCCUGCCGUUCCUGUUAUUGUUUUGUUGUUAAUUGUCAGCGAUUAAACGGCGUAGACAACAACAAAUAACGAU ...((((((...((((-......))))....))))))((((..((((.((((((((((((((((((....))))).))))))).)))))).))))...)))).. ( -30.70) >DroSec_CAF1 27998 103 + 1 UUCGCGGGCUUCUCUC-AAAGCGAAGGCAGUGCCUGCCGUUCCUGUUUUUGUUUUGCUGUUAAUUGUCAGCGAUUAAACGGCGUAGACAACAACAAAUAACGAU ...((((((....((.-...((....)))).))))))((((..((((.((((((((((((((((((....))))).))))))).)))))).))))...)))).. ( -34.20) >DroSim_CAF1 33750 103 + 1 UUCGCGGGCUUCUCUC-AAAGCAAAGGCAGUGCCUGCCGUUCCUGUUUUUGUUUUGCUGUUAAUUGUCAGCGAUUAAACGGCGUAGACAACAACAAAUAACGAU ...((((((..((.((-........)).)).))))))((((..((((.((((((((((((((((((....))))).))))))).)))))).))))...)))).. ( -32.80) >DroEre_CAF1 29226 101 + 1 GUCGCGGUCUUCUCUCAAAAGCGAAGGCAGUGCCUGCCGUUCU---UUUUGUUUUGUUGUUAAUUGUCAGCGAUUGAACGGCGGAGACAACAACAAAUAACGAU (((((.((((((.((....)).))))))...))(((((((((.---.......((((((........))))))..))))))))).)))................ ( -31.10) >DroYak_CAF1 29590 100 + 1 GUCGCGGUUUUCUCUG-AAAGCGAAGGCAGUGCCUGCCGUUCC---UUUUGUUUUGUUGUUAAUUGUCAGCGAUUGAACGGCGUAGACAACAACAAAUAACGAU ((((..((((.....(-((((.((((((((...))))).))))---))))((((((((((((((((....)))))).)))))).))))......))))..)))) ( -26.40) >consensus UUCGCGGGCUUCUCUC_AAAGCGAAGGCAGUGCCUGCCGUUCCUGUUUUUGUUUUGUUGUUAAUUGUCAGCGAUUAAACGGCGUAGACAACAACAAAUAACGAU ...((((((..((.......((....)))).))))))((((..((((.((((((((((((((((((....)))))).)))))).)))))).))))...)))).. (-25.39 = -26.19 + 0.80)

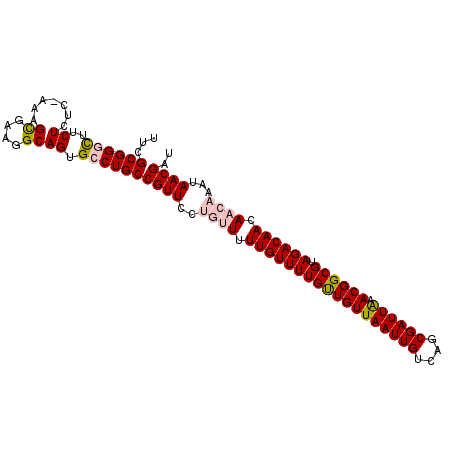
| Location | 19,715,620 – 19,715,723 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 92.53 |
| Mean single sequence MFE | -25.44 |
| Consensus MFE | -21.78 |
| Energy contribution | -21.38 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.86 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.527375 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19715620 103 - 27905053 AUCGUUAUUUGUUGUUGUCUACGCCGUUUAAUCGCUGACAAUUAACAACAAAACAAUAACAGGAACGGCAGGCACUGCCUUCACUUU-AAGAGAAGCCCGCGAA .((((..((((((((((((..((.........))..))))....)))))))).........((...(((((...)))))....((((-....)))).)))))). ( -22.20) >DroSec_CAF1 27998 103 - 1 AUCGUUAUUUGUUGUUGUCUACGCCGUUUAAUCGCUGACAAUUAACAGCAAAACAAAAACAGGAACGGCAGGCACUGCCUUCGCUUU-GAGAGAAGCCCGCGAA .((((..((((((((((((..((.........))..))))....)))))))).........((...(((((...)))))...(((((-....))))))))))). ( -28.60) >DroSim_CAF1 33750 103 - 1 AUCGUUAUUUGUUGUUGUCUACGCCGUUUAAUCGCUGACAAUUAACAGCAAAACAAAAACAGGAACGGCAGGCACUGCCUUUGCUUU-GAGAGAAGCCCGCGAA .((((..((((((((((((..((.........))..))))....)))))))).........((...(((((...)))))...(((((-....))))))))))). ( -28.30) >DroEre_CAF1 29226 101 - 1 AUCGUUAUUUGUUGUUGUCUCCGCCGUUCAAUCGCUGACAAUUAACAACAAAACAAAA---AGAACGGCAGGCACUGCCUUCGCUUUUGAGAGAAGACCGCGAC .((((..((((((((((..((.((.(......))).))....)))))))))).(((((---.(((.(((((...))))))))..)))))..........)))). ( -24.90) >DroYak_CAF1 29590 100 - 1 AUCGUUAUUUGUUGUUGUCUACGCCGUUCAAUCGCUGACAAUUAACAACAAAACAAAA---GGAACGGCAGGCACUGCCUUCGCUUU-CAGAGAAAACCGCGAC .((((..((((((((((((..((.........))..))))....))))))))...(((---((((.(((((...)))))))).))))-...........)))). ( -23.20) >consensus AUCGUUAUUUGUUGUUGUCUACGCCGUUUAAUCGCUGACAAUUAACAACAAAACAAAAACAGGAACGGCAGGCACUGCCUUCGCUUU_GAGAGAAGCCCGCGAA .((((..((((((((((((..((.........))..))))....))))))))..........(((.(((((...)))))))).................)))). (-21.78 = -21.38 + -0.40)

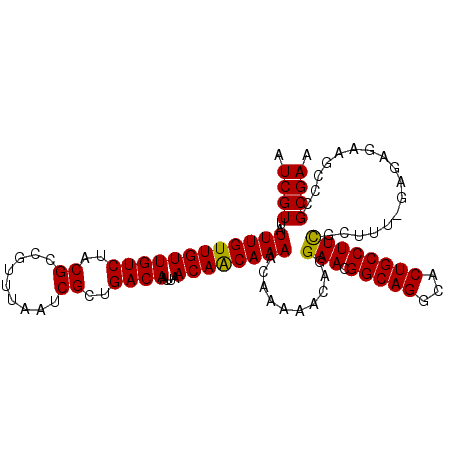

| Location | 19,715,654 – 19,715,763 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 94.94 |
| Mean single sequence MFE | -33.08 |
| Consensus MFE | -30.46 |
| Energy contribution | -31.18 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.95 |
| SVM RNA-class probability | 0.983766 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19715654 109 + 27905053 GCCGUUCCUGUUAUUGUUUUGUUGUUAAUUGUCAGCGAUUAAACGGCGUAGACAACAACAAAUAACGAUGCGUGGUUAUGCGGGCGGUGGUUUUUGUUCGUUGGAGCAA (((((((.((((.((((((((((((((((((....))))).))))))).)))))).)))).........((((....))))))))))).....((((((....)))))) ( -32.30) >DroSec_CAF1 28032 109 + 1 GCCGUUCCUGUUUUUGUUUUGCUGUUAAUUGUCAGCGAUUAAACGGCGUAGACAACAACAAAUAACGAUGCGUGGUUAUGCGGGCGGUGGUUUUUGUUCGUUGGAGCAA (((((((.((((.((((((((((((((((((....))))).))))))).)))))).)))).........((((....))))))))))).....((((((....)))))) ( -34.80) >DroSim_CAF1 33784 109 + 1 GCCGUUCCUGUUUUUGUUUUGCUGUUAAUUGUCAGCGAUUAAACGGCGUAGACAACAACAAAUAACGAUGCGUGGUUAUGCGGGCGGUGGUUUUUGUUCGUUGGAGCAA (((((((.((((.((((((((((((((((((....))))).))))))).)))))).)))).........((((....))))))))))).....((((((....)))))) ( -34.80) >DroEre_CAF1 29261 106 + 1 GCCGUUCU---UUUUGUUUUGUUGUUAAUUGUCAGCGAUUGAACGGCGGAGACAACAACAAAUAACGAUGCGUGAUGAUGCGGGCGGUGGUUUUUGUUCGUUGGAGCAA (((((((.---..((((((((((((((((((....))))).......(....)))))))))..))))).((((....))))))))))).....((((((....)))))) ( -30.00) >DroYak_CAF1 29624 106 + 1 GCCGUUCC---UUUUGUUUUGUUGUUAAUUGUCAGCGAUUGAACGGCGUAGACAACAACAAAUAACGAUGCGUGCUGAUGCGGGCGGUGGUUUUUGUUCGUUGGAGCAA (((((((.---.......((((((........))))))..)))))))((...((((((((((.(((.((((.(((....))).)).)).))))))))).))))..)).. ( -33.50) >consensus GCCGUUCCUGUUUUUGUUUUGUUGUUAAUUGUCAGCGAUUAAACGGCGUAGACAACAACAAAUAACGAUGCGUGGUUAUGCGGGCGGUGGUUUUUGUUCGUUGGAGCAA (((((((.((((.((((((((((((((((((....)))))).)))))).)))))).)))).........((((....))))))))))).....((((((....)))))) (-30.46 = -31.18 + 0.72)

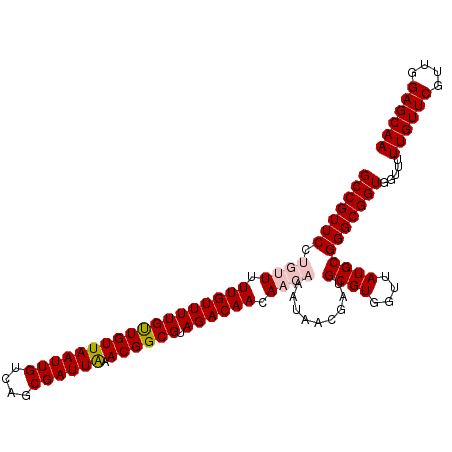

| Location | 19,715,723 – 19,715,842 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.65 |
| Mean single sequence MFE | -37.92 |
| Consensus MFE | -30.72 |
| Energy contribution | -31.32 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.544325 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19715723 119 + 27905053 GCGUGGUUAUGCGGGCGGUGGUUUUUGUUCGUUGGAGCAAGUGUGACUGCAGGUCGCGAGGCUAGCGAGCCCACUCGAAAUGGCUACACCAAAAAGUGUGACUAAAAAAUACCAAA-AUA ...((((((((((((((((.(..(((((((....)))))))..).)))((.((((....)))).))..))))........(((.....)))....)))))))))............-... ( -38.20) >DroSec_CAF1 28101 119 + 1 GCGUGGUUAUGCGGGCGGUGGUUUUUGUUCGUUGGAGCAAGUGUGACUGCAGGUCGCGAGGCUAGCGAGCCCACUCGAAAUGGCUACACCGAAAAGUGUGACUAAAAAAUACCAAA-AUA ...(((((((((...(((((...(((((((....)))))))((((((.....)))))).(((((.((((....))))...))))).)))))....)))))))))............-... ( -40.30) >DroSim_CAF1 33853 119 + 1 GCGUGGUUAUGCGGGCGGUGGUUUUUGUUCGUUGGAGCAAGUGUGACUGCAGGUCGCGAGGCUAGCGAGCCCACUCGAAAUGGCUACACCGAAAAGUGUGACUAAAAAAUACCAAA-AUA ...(((((((((...(((((...(((((((....)))))))((((((.....)))))).(((((.((((....))))...))))).)))))....)))))))))............-... ( -40.30) >DroEre_CAF1 29327 119 + 1 GCGUGAUGAUGCGGGCGGUGGUUUUUGUUCGUUGGAGCAAGUGUGACUGCAGGUCGCGAAACUGGAGAGCUCACUCGAUGUGGCUGCACAGA-AAGUAUGACUAAAAAAUACCAAACAAA ((((....))))(((((((.(..(((((((....)))))))..).))))).(((((.....(((...(((.(((.....))))))...))).-.....)))))........))....... ( -33.80) >DroYak_CAF1 29690 120 + 1 GCGUGCUGAUGCGGGCGGUGGUUUUUGUUCGUUGGAGCAAGUGUGACUGCAGGUCGCGAAACUGGAGAGCCCACUCGAAACGCCUACACUGAAAAGUGUGCCUAAAAAAUACCAAGCAAA ((((..(((.(.((((..((((((((((((....)))))...(((((.....))))))))))))....)))).))))..)))).((((((....)))))).................... ( -37.00) >consensus GCGUGGUUAUGCGGGCGGUGGUUUUUGUUCGUUGGAGCAAGUGUGACUGCAGGUCGCGAGGCUAGCGAGCCCACUCGAAAUGGCUACACCGAAAAGUGUGACUAAAAAAUACCAAA_AUA ((((....))))(((((((.(..(((((((....)))))))..).))))).((((....(((((.((((....))))...)))))((((......))))))))........))....... (-30.72 = -31.32 + 0.60)

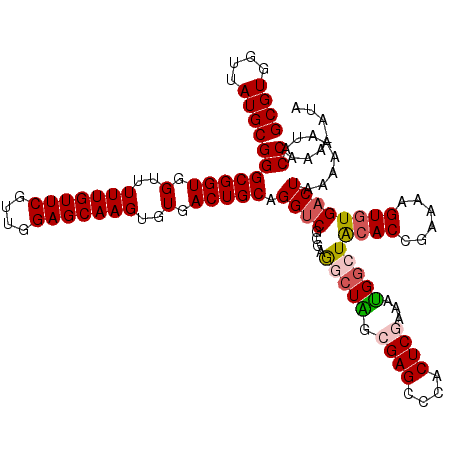
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:45:16 2006